Gromos Tutorial¶
This tutorial will provide a quick introduction to how to setup, perform and analyze a MD simulation of a peptide in Gromos.
First we will generated the required input files, afterwards the simulation will be prepared with energy minimizations and equilibration. Finally a short MD simulation will be executed and quickly analyzed.
But first some minor definitions will be prepared.
[1]:
import os
# Check if the path to this pacakge is set, else try to add it.
try:
import pygromos
except:
import os, sys, copy
root_dir = os.getcwd()
sys.path.append(root_dir+"/..")
#if package is not installed and path not set correct - this helps you out :)
import pygromos
from pygromos.utils import bash
[2]:
#General path definitions:
## Project dir - The project dir will contain all files and results generated from this notebook.
project_dir = os.path.abspath("example_files/Tutorial_System")
input_dir = project_dir+"/input" # contains prepared files (pdb of the peptide)
## Gromos Bin Path
gromosXX_bin = None
gromosPP_bin = None #None uses the Path variable - can be used . if you require a specific compiled gromos version, add a path here.
Build the input files¶
When a simulation study of a particular system or process is to be carried out, a number of choices have to be made with respect to the set-up of the simulation.
In PyGromosTools, the central hub to manage all these files is a GromosSystem object. Each file can be stored or generated with this object and it can be used to construct the system, that should be simulated.
The essential files of a Simulation are: * a topology file, containing all the topological and force-field information of the molecular system to be studied. * a coordinate file, representing the system of interest. * a simulation parameter file, telling the simulation engine, which simulation technique should be used and which physical parameters should be set.
[3]:
from pygromos.files.gromos_system import Gromos_System
# Build Gromos System object
build_system = Gromos_System(work_folder=project_dir, system_name='peptide')
#set file building folder
build_system.work_folder = bash.make_folder(project_dir+"/a_build_initial_files")
/home/mlehner/PyGromosTools/pygromos/files/gromos_system/gromos_system.py:869: UserWarning: Did not change file path as its only promised None
warnings.warn("Did not change file path as its only promised " + str(file_obj.path))
Expert Tip: Whenever you wonder about what a class or a function in our package does, you can use the standard help function of Python to get the information of the Docstring of the source code! This often helps understanding the code.
[4]:
help(build_system)
Help on Gromos_System in module pygromos.files.gromos_system.gromos_system object:
class Gromos_System(builtins.object)
| Gromos_System(work_folder: str, system_name: str, rdkitMol: rdkit.Chem.rdchem.Mol = None, in_mol2_file: str = None, readIn: bool = True, forcefield: pygromos.files.forcefield._generic_force_field._generic_force_field = <pygromos.files.forcefield._generic_force_field._generic_force_field object at 0x7f799f5a9460>, auto_convert: bool = False, adapt_imd_automatically: bool = True, verbose: bool = False, in_smiles: str = None, in_residue_list: List = None, in_top_path: str = None, in_cnf_path: str = None, in_imd_path: str = None, in_disres_path: str = None, in_ptp_path: str = None, in_posres_path: str = None, in_refpos_path: str = None, in_qmmm_path: str = None, in_gromosXX_bin_dir: str = None, in_gromosPP_bin_dir: str = None)
|
| Methods defined here:
|
| __deepcopy__(self, memo)
|
| __getstate__(self)
| preperation for pickling:
| remove the non trivial pickling parts
|
| __init__(self, work_folder: str, system_name: str, rdkitMol: rdkit.Chem.rdchem.Mol = None, in_mol2_file: str = None, readIn: bool = True, forcefield: pygromos.files.forcefield._generic_force_field._generic_force_field = <pygromos.files.forcefield._generic_force_field._generic_force_field object at 0x7f799f5a9460>, auto_convert: bool = False, adapt_imd_automatically: bool = True, verbose: bool = False, in_smiles: str = None, in_residue_list: List = None, in_top_path: str = None, in_cnf_path: str = None, in_imd_path: str = None, in_disres_path: str = None, in_ptp_path: str = None, in_posres_path: str = None, in_refpos_path: str = None, in_qmmm_path: str = None, in_gromosXX_bin_dir: str = None, in_gromosPP_bin_dir: str = None)
| The Gromos_System class is the central unit of PyGromosTools for files and states.
| With this class all files can be read-in or the files can be automatically generated from smiles.
| Additionally to that can all gromos++ functions be used from the Gromos System, so system generation can be easily accomplished.
|
| if you want to remove all binary checks, do the following:
| >>> from pygromos.files.gromos_system.gromos_system import Gromos_System
| >>> Gromos_System._gromos_noBinary_checks = True
| >>> sys = Gromos_System()
|
| Parameters
| ----------
| work_folder : str
| This gives the initial working folder for the system.
| system_name : str
| the name of the system, also used as file prefix
| in_smiles : str, optional
| Molecule input SMILES for file generation, by default None
| in_top_path : str, optional
| input Gromos topology path (.top), by default None
| in_cnf_path : str, optional
| input Gromos coordinate path (.cnf), by default None
| in_imd_path : str, optional
| input Gromos simulation parameters path (.imd), by default None
| in_disres_path : str, optional
| input Gromos distance restraint path (.disres), by default None
| in_ptp_path : str, optional
| input pertubation file for free energy calculations (.ptp), by default None
| in_posres_path : str, optional
| input position restraints file (.por), by default None
| in_qmmm_path: str, optional
| qmmm parameter file (.qmmm), by default None
| in_refpos_path : str, optional
| input reference position file (.rpf), by default None
| in_gromosXX_bin_dir : str, optional
| path to the binary dir of GromosXX, by default None -> uses the set binaries in the PATH variable
| in_gromosPP_bin_dir : str, optional
| path to the binary dir of GromosPP, by default None -> uses the set binaries in the PATH variable
| rdkitMol : Chem.rdchem.Mol, optional
| input rdkit Molecule, by default None
| in_mol2_file : str, optional
| path to input mol2 file, by default None
| readIn : bool, optional
| readIn all provided files?, by default True
| Forcefield : forcefield_system, optional
| input PyGromos - forcefield Class , by default forcefield_system()
| auto_convert : bool, optional
| automatically convert rdkit MOL and smiles to gromos files, by default False
| adapt_imd_automatically : bool, optional
| adjust the input imd file to the GromosSystem, by default True
| verbose : bool, optional
| Stay a while and listen!, by default False
|
| Raises
| ------
| Warning
| Rises warning if files are not present.
|
| __repr__(self) -> str
| Return repr(self).
|
| __setstate__(self, state)
|
| __str__(self) -> str
| Return str(self).
|
| adapt_imd(self, not_ligand_residues: List[str] = [])
|
| auto_convert(self)
|
| copy(self)
|
| generate_posres(self, residues: List = [<class 'int'>], keep_residues: bool = True, verbose: bool = False)
|
| get_file_paths(self) -> Dict[str, str]
| get the paths of the files in a dict.
| Returns
| -------
| Dict[str, str]
| returns alle file paths, with attribute file name as key.
|
| get_script_generation_command(self, var_name: str = None, var_prefixes: str = '') -> str
|
| parse_attribute_files(self, file_mapping: Dict[str, str], readIn: bool = True, verbose: bool = False)
| This function sets dynamically builds the output folder, the file objs of this class and checks their dependencies.
|
| Parameters
| ----------
| file_mapping: Dict[str, Union[str, None]]
| attribute name: input path
|
| Returns
| -------
|
| prepare_for_simulation(self, not_ligand_residues: List[str] = [])
|
| rdkit2Gromos(self)
|
| rdkit2GromosName(self) -> str
|
| rdkitImport(self, inputMol: rdkit.Chem.rdchem.Mol)
|
| read_files(self, verbose: bool = False)
|
| rebase_files(self)
|
| save(self, path: Union[str, _io.FileIO] = None, safe: bool = True) -> str
| This method stores the Class as binary obj to a given path or fileBuffer.
|
| work_folder_no_update(self, work_folder: str)
| # Updates the work folder without updating all file paths
|
| write_files(self, cnf: bool = False, imd: bool = False, top: bool = False, ptp: bool = False, disres: bool = False, posres: bool = False, refpos: bool = False, qmmm: bool = False, mol: bool = False, all: bool = True, verbose: bool = False)
|
| ----------------------------------------------------------------------
| Class methods defined here:
|
| load(path: Union[str, _io.FileIO] = None) -> object from builtins.type
| This method stores the Class as binary obj to a given path or fileBuffer.
|
| ----------------------------------------------------------------------
| Readonly properties defined here:
|
| all_file_paths
| Returns
| -------
|
| all_files
| Returns
| -------
|
| ----------------------------------------------------------------------
| Data descriptors defined here:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
|
| cnf
|
| disres
|
| gromosPP
|
| gromosPP_bin_dir
|
| gromosXX
|
| gromosXX_bin_dir
|
| imd
|
| name
|
| posres
|
| ptp
|
| qmmm
|
| refpos
|
| top
|
| trc
|
| tre
|
| work_folder
|
| ----------------------------------------------------------------------
| Data and other attributes defined here:
|
| __annotations__ = {'_checkpoint_path': <class 'str'>, '_future_promise...
|
| optional_files = {'disres': <class 'pygromos.files.topology.disres.Dis...
|
| required_files = {'cnf': <class 'pygromos.files.coord.cnf.Cnf'>, 'imd'...
|
| traj_files = {'trc': <class 'pygromos.files.trajectory.trc.Trc'>, 'tre...
The Topology File¶
The first task is to generate a molecular topology file containing the topological and force-field data concerning the molecular system under consideration. Specifying a complete molecular topology for a large molecule, however, is a tedious task. Therefore, in GROMOS a molecular topology is generated from molecular topology building blocks which carry the topological information of molecules like amino acid residues, nucleotides, etc., see Vol. 3. The molecular topology building blocks can be linked together into a complete molecular topology file using the GROMOS++ program make_top.
Many molecular topology building blocks are available in the molecular topology building block files (*.mtb), which are standard data files that come together with the GROMOS package.
In case the needed molecular topology building blocks are not part of the standard distribution, they must be constructed. Constructing a new topology building block may require estimation of additional force-field parameters, which have to be added to the interaction function parameter file (*.ifp).
When generating a molecular topology for the system of interest one should also address the protonation state of the molecular groups according to the pH and of the solvent and counter ions that need to be included in the simulation box. In case of a molecular complex, e.g. a DNA-protein complex, the two separately generated molecular topologies for the protein and the DNA can be merged using the GROMOS++ program com top.
Building the Topology File¶
In this section you should build a molecular topology of a linear charged penta-peptide with water as a solvent, including two Cl− counter ions.
programs: make_top, check_top
build topology for single molecule¶
You will build the molecular topology file of the linear charged penta-peptide Val-Tyr-Arg-Lys-Gln by using the GROMOS++ program make_top. As input following parameters will be provided: * in_building_block_lib_path: the molecular topology building block file is specified. In the code below, you will see, that we gather the forcefield information from the Gromos54A7 force field, which is directly provided by PyGromosTools * in_parameter_lib_path: specifies the interaction function parameter file. Also here we take the parameters directly from the Gromos54A7 forcefield from PyGromosTools. * in_solvent: The string name of the desired solvent. * in_sequence: the sequence of the building blocks for the amino acid residues, including the amino and carboxy terminus is specified (NH3+ VAL TYR ARG LYSH GLN COO-). (Notice that both termini are charged)
[5]:
from pygromos.data.ff import Gromos54A7 #Get standard information of the GROMOS54A7 force field.
#Generate the topology
build_system.make_top(in_sequence="NH3+ VAL TYR ARG LYSH GLN COO-",
in_solvent="H2O",
in_building_block_lib_path = Gromos54A7.mtb,
in_parameter_lib_path= Gromos54A7.ifp)
#Here the residues in the topology file will be printed out :) - this corresponds to the RESNAME Block in the topology file:
build_system.top.RESNAME
[5]:
RESNAME
5
VAL
TYR
ARG
LYSH
GLN
END
The Coordinate File¶
Coordinates for biomolecules are often available from X-ray or NMR experiments and can be obtained in Protein Data Bank (PDB) format, which can be converted to GROMOS format using the GROMOS++ program pdb2g96. However, the conversion is not always straightforward since the naming and numbering of the atoms in the PDB format usually do not match the GROMOS format.
Moreover, the coordinates for hydrogen atoms are not present in the PDB files (when the structure was determined using X-ray diffraction data) and have to be generated using the GROMOS++ program gch.
When the structure is determined using NMR data, the PDB structure often contains more hydrogen atoms than are needed for GROMOS, as in the GROMOS force field only polar and aromatic hydrogens are explicitly represented. Aliphatic hydrogens are non-existing due to the use of so-called united atoms. The aliphatic hydrogen and carbon atoms are merged to form united atoms which have their own parameters. If no atomic coordinates for the solute are available from experimental data, the coordinates have to be generated using molecular modeling software. Often parts of the structure (e.g. flexible loops) are not resolved in the experiment and therefore not available in the PDB and have to be modeled as well.
Periodic Boxes¶
When a simulation of a solute in solution is to be carried out, a (periodic) box (be it rectangular, triclinic or truncated octahedral) is put around the solute and filled with solvent molecules up to the required density. The solvent coordinates can e.g. be generated using the GROMOS++ program sim box. The generated box should be sufficiently large to allow the use of a reasonable non-bonded interaction cut-off radius. Putting the solute in a box of solvent using the sim_box program will result in several high-energy atom-atom contacts at the solute-solvent interface and at the box edges. In order to relax the generated configuration the solvent configuration should be energy minimized while positionally restraining the solute. Counter-ion atomic coordinates can then be generated using the GROMOS++ program ion, which can replace a number of solvent molecules by ions.
programs: pdb2g96, gch
Building the Coordinate File¶
The initial coordinate .pdb file is already provided in the input directory as peptide.pdb. Open the file peptide.pdb and check if the atom names match the names in the molecular topology object SOLUTEATOM block. In the pdb file peptide.pdb the coordinates for hydrogen atoms are not given and have to be generated. If the atom names are correct, you can continue in the second next cell by converting the PDB file peptide.pdb into the GROMOS format using the GROMOS++ program pdb2g96. The hydrogen atoms will be added to the coordinate file according to the topological requirements.
Warning: When converting coordinate files from the Protein Data Bank to GROMOS format many difficulties may emerge. If you encounter problems using the pdb2g96 program, have a look at Sec. 4-7.3. There you can find further documentation on the advanced usage of this program. Especially the use of a library that matches residue and atom names might be useful in many cases. pdb2g96.lib which you can find in the directory is an example of the PDB library file. ***
[6]:
#Compare the atom names of the pdb file input/peptide.pdb with the printed ones here:
build_system.top.SOLUTEATOM
[6]:
SOLUTEATOM
# NRP: number of solute atoms
71
# ATNM: atom number
# MRES: residue number
# PANM: atom name of solute atom
# IAC: integer (van der Waals) atom type code
# MASS: mass of solute atom
# CG: charge of solute atom
# CGC: charge group code (0 or 1)
# INE: number of excluded atoms
# INE14: number of 1-4 interactions
# ATNM MRES PANM IAC MASS CG CGC INE
# INE14
1 1 H1 21 1.008 0.248 0 4 2 3 4 5
2 6 9
2 1 H2 21 1.008 0.248 0 3 3 4 5
2 6 9
3 1 N 8 14.0067 0.129 0 4 4 5 6 9
4 7 8 10 11
4 1 H3 21 1.008 0.248 0 1 5
2 6 9
5 1 CA 14 13.019 0.127 1 6 6 7 8 9 10 11
2 12 13
6 1 CB 14 13.019 0.0 0 3 7 8 9
2 10 11
7 1 CG1 16 15.035 0.0 0 1 8
1 9
8 1 CG2 16 15.035 0.0 1 0
1 9
9 1 C 12 12.011 0.45 0 4 10 11 12 13
2 14 27
10 1 O 1 15.9994 -0.45 1 1 11
2 12 13
11 2 N 6 14.0067 -0.31 0 4 12 13 14 27
3 15 28 29
12 2 H 21 1.008 0.31 1 1 13
2 14 27
13 2 CA 14 13.019 0.0 0 5 14 15 27 28 29
4 16 18 30 31
14 2 CB 15 14.027 0.0 0 8 15 16 17 18 19 20
22 27
2 28 29
15 2 CG 12 12.011 0.0 1 9 16 17 18 19 20 21
22 23 24
1 27
16 2 CD1 12 12.011 -0.14 0 8 17 18 19 20 21 22
24 25
0
17 2 HD1 20 1.008 0.14 1 4 18 20 21 24
0
18 2 CD2 12 12.011 -0.14 0 6 19 20 22 23 24 25
0
19 2 HD2 20 1.008 0.14 1 3 22 23 24
0
20 2 CE1 12 12.011 -0.14 0 5 21 22 23 24 25
1 26
21 2 HE1 20 1.008 0.14 1 3 22 24 25
0
22 2 CE2 12 12.011 -0.14 0 3 23 24 25
1 26
23 2 HE2 20 1.008 0.14 1 2 24 25
0
24 2 CZ 12 12.011 0.203 0 2 25 26
0
25 2 OH 3 15.9994 -0.611 0 1 26
0
26 2 HH 21 1.008 0.408 1 0
0
27 2 C 12 12.011 0.45 0 4 28 29 30 31
2 32 44
28 2 O 1 15.9994 -0.45 1 1 29
2 30 31
29 3 N 6 14.0067 -0.31 0 4 30 31 32 44
3 33 45 46
30 3 H 21 1.008 0.31 1 1 31
2 32 44
31 3 CA 14 13.019 0.0 0 5 32 33 44 45 46
3 34 47 48
32 3 CB 15 14.027 0.0 0 3 33 34 44
3 35 45 46
33 3 CG 15 14.027 0.0 1 2 34 35
3 36 37 44
34 3 CD 15 14.027 0.09 0 3 35 36 37
2 38 41
35 3 NE 11 14.0067 -0.11 0 4 36 37 38 41
4 39 40 42 43
36 3 HE 21 1.008 0.24 0 1 37
2 38 41
37 3 CZ 12 12.011 0.34 0 6 38 39 40 41 42 43
0
38 3 NH1 10 14.0067 -0.26 0 3 39 40 41
2 42 43
39 3 HH11 21 1.008 0.24 0 1 40
1 41
40 3 HH12 21 1.008 0.24 0 0
1 41
41 3 NH2 10 14.0067 -0.26 0 2 42 43
0
42 3 HH21 21 1.008 0.24 0 1 43
0
43 3 HH22 21 1.008 0.24 1 0
0
44 3 C 12 12.011 0.45 0 4 45 46 47 48
2 49 57
45 3 O 1 15.9994 -0.45 1 1 46
2 47 48
46 4 N 6 14.0067 -0.31 0 4 47 48 49 57
3 50 58 59
47 4 H 21 1.008 0.31 1 1 48
2 49 57
48 4 CA 14 13.019 0.0 0 5 49 50 57 58 59
3 51 60 61
49 4 CB 15 14.027 0.0 1 3 50 51 57
3 52 58 59
50 4 CG 15 14.027 0.0 0 2 51 52
2 53 57
51 4 CD 15 14.027 0.0 1 2 52 53
3 54 55 56
52 4 CE 15 14.027 0.127 0 4 53 54 55 56
0
53 4 NZ 8 14.0067 0.129 0 3 54 55 56
0
54 4 HZ1 21 1.008 0.248 0 2 55 56
0
55 4 HZ2 21 1.008 0.248 0 1 56
0
56 4 HZ3 21 1.008 0.248 1 0
0
57 4 C 12 12.011 0.45 0 4 58 59 60 61
2 62 69
58 4 O 1 15.9994 -0.45 1 1 59
2 60 61
59 5 N 6 14.0067 -0.31 0 4 60 61 62 69
3 63 70 71
60 5 H 21 1.008 0.31 1 1 61
2 62 69
61 5 CA 14 13.019 0.0 0 5 62 63 69 70 71
1 64
62 5 CB 15 14.027 0.0 0 3 63 64 69
4 65 66 70 71
63 5 CG 15 14.027 0.0 1 3 64 65 66
3 67 68 69
64 5 CD 12 12.011 0.29 0 4 65 66 67 68
0
65 5 OE1 1 15.9994 -0.45 0 1 66
2 67 68
66 5 NE2 7 14.0067 -0.72 0 2 67 68
0
67 5 HE21 21 1.008 0.44 0 1 68
0
68 5 HE22 21 1.008 0.44 1 0
0
69 5 C 12 12.011 0.27 0 2 70 71
0
70 5 O1 2 15.9994 -0.635 0 1 71
0
71 5 O2 2 15.9994 -0.635 1 0
0
END
[7]:
# Generate coordinate file:
build_system.pdb2gromos(in_pdb_path=input_dir+"/peptide.pdb")
#show the coordinates that were generated.
build_system.cnf.view
Initial Write out¶
Next we will write out all generated files, such we have the initial products of our efforts.
[8]:
# Now write all files, such that you can check them directly.
print("Path before rebase: "+str(build_system.cnf.path))
build_system.rebase_files()
#check this
print("Path after rebase: "+build_system.cnf.path)
#Check also how the system path and attributes were automatically updated.
build_system
Path before rebase: None
File imd is empty , can not be written!
Path after rebase: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/a_build_initial_files/peptide.cnf
[8]:
GROMOS SYSTEM: peptide
################################################################################
WORKDIR: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/a_build_initial_files
LAST CHECKPOINT: None
GromosXX_bin: None
GromosPP_bin: None
FILES:
imd: None
top: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/a_build_initial_files/peptide.top
cnf: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/a_build_initial_files/peptide.cnf
FUTURE PROMISE: False
SYSTEM:
PROTEIN: protein nresidues: 5 natoms: 71
LIGANDS: [] resID: [] natoms: 0
Non-LIGANDS: [] nmolecules: 0 natoms: 0
SOLVENT: [] nmolecules: 0 natoms: 0
add hydrogens¶
Have a look at the cnf coordinates in the GromosSystem (next cell). You will notice that the hydrogen atoms have been added to the coordinate file with the Cartesian coordinates being set to zero.
[9]:
#print x lines of the Position Block of valine
print("".join([str(atomP) for atomP in build_system.cnf.POSITION if("VAL" == atomP.resName)]))
1 VAL H1 1 0.000000000 0.000000000 0.000000000
1 VAL H2 2 0.000000000 0.000000000 0.000000000
1 VAL N 3 1.196200000 1.413300000 1.529800000
1 VAL H3 4 0.000000000 0.000000000 0.000000000
1 VAL CA 5 1.237300000 1.322400000 1.421900000
1 VAL CB 6 1.116100000 1.268700000 1.345400000
1 VAL CG1 7 1.041700000 1.369500000 1.257600000
1 VAL CG2 8 1.157800000 1.148100000 1.261100000
1 VAL C 9 1.335700000 1.393300000 1.328600000
1 VAL O 10 1.447800000 1.344200000 1.316600000
In order to generate meaningful coordinates for the hydrogen atoms run the GROMOS++ program protonate (or gch). It will generate the coordinates for hydrogen atoms by geometric means using the information from the molecular topology file. The argument tolerance sets the tolerance that is used for keeping the coordinates of hydrogens that are already present in the coordinate file.
[10]:
# Add the hydrogen positions
build_system.add_hydrogens()
#store the current files with a different name:
build_system.name = "peptideH"
build_system.rebase_files()
#visualize again the nice structure
build_system.cnf.view
File imd is empty , can not be written!
Optional: Convert cnf to pdb¶
[11]:
pdb_path = build_system.cnf.write_pdb(build_system.work_folder+"/vacuum_hpeptide.pdb")
pdb_path
[11]:
'/home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/a_build_initial_files/vacuum_hpeptide.pdb'
Energy Minimization - Vacuum¶
Before putting the penta-peptide into a box of solvent, its configuration can be relaxed by energy minimisation.
Simulation Parameter File¶
The GROMOS simulation parameter file (also called imd) template_emin_vac can be parsed like follows and contains the following blocks:
[12]:
from pygromos.data.simulation_parameters_templates import template_emin_vac
#load simulation parameter file (Imd) File
build_system.imd = template_emin_vac
#for nicer code we will store the simulation parameter file in a variable.
emin_vac_imd_file = build_system.imd
TITLE Block¶
In the TITLE block you specify what is done with following input file, so you know what you did with it later and can easily reuse it.
[13]:
emin_vac_imd_file.TITLE
[13]:
TITLE
steepest descent energy minimization of the peptide in vacuum
>>> Generated with PyGromosTools (riniker group) <<<
END
ENERGYMIN Block¶
The existence of the ENERGYMIN block means that Gromos will perform an energy minimisation (EM) run. The NTEM switch indicates which minimisation algorithm to be used. With NTEM = 1 we indicate that the steepest-descent algorithm (Sec. 2-11.2) is used. NCYC gives the number of steps before resetting of conjugate-gradient search direction in case we would use the conjugate gradient method (NTEM = 2). Using DELE the energy threshold (the difference in energy between two energy minimisation steps) for stopping the minimisation process (convergence) is specified. The initial step size and maximum step size is given in DX0 and DXM, respectively. Using FLIM the absolute value of the forces can be limited to a maximum value before the algorithm is applied (see also 4-93).
[14]:
help(emin_vac_imd_file.ENERGYMIN)
Help on ENERGYMIN in module pygromos.files.blocks.imd_blocks object:
class ENERGYMIN(_generic_imd_block)
| ENERGYMIN(NTEM: int = 0, NCYC: int = 0, DELE: float = 0, DX0: float = 0, DXM: float = 0, NMIN: int = 0, FLIM: float = 0, content: List[str] = None)
|
| ENERGYMIN block
| This block takes care of managing the Energyminimization controls.
|
| Attributes
| ----------
| NTEM: int
| controls energy minimisation mode.
| 0: do not do energy minimisation (default)
| 1: steepest-descent minimisation
| 2: Fletcher-Reeves conjugate-gradient minimisation
| 3: Polak-Ribiere conjugate-gradient minimisation
| NCYC: int
| >0 number of steps before resetting the conjugate-gradient search direction
| =0 reset only if the energy grows in the search direction
| DELE: float
| >0.0 energy threshold for convergence
| >0.0 (conjugate-gradient) RMS force threshold for convergence
| DX0: float
| >0.0 initial step size
| DXM: float
| >0.0 maximum step size
| NMIN: float
| >0 minimum number of minimisation steps
| FLIM: float
| >=0.0 limit force to maximum value (FLIM > 0.0 is not recommended)
|
| Method resolution order:
| ENERGYMIN
| _generic_imd_block
| pygromos.files.blocks._general_blocks._generic_gromos_block
| builtins.object
|
| Methods defined here:
|
| __init__(self, NTEM: int = 0, NCYC: int = 0, DELE: float = 0, DX0: float = 0, DXM: float = 0, NMIN: int = 0, FLIM: float = 0, content: List[str] = None)
| Initialize self. See help(type(self)) for accurate signature.
|
| ----------------------------------------------------------------------
| Data and other attributes defined here:
|
| __annotations__ = {'DELE': <class 'float'>, 'DX0': <class 'float'>, 'D...
|
| name = 'ENERGYMIN'
|
| ----------------------------------------------------------------------
| Methods inherited from _generic_imd_block:
|
| block_to_string(self) -> str
|
| read_content_from_str(self, content: List[str])
|
| ----------------------------------------------------------------------
| Methods inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __copy__(self)
|
| __deepcopy__(self, memo)
|
| __eq__(self, _generic_gromos_block__o: object) -> bool
| Return self==value.
|
| __iter__(self)
|
| __repr__(self)
| Return repr(self).
|
| __str__(self)
| Return str(self).
|
| get_name(self)
|
| ----------------------------------------------------------------------
| Data descriptors inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
|
| ----------------------------------------------------------------------
| Data and other attributes inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __hash__ = None
[15]:
# The current ENERGYMIN Block
emin_vac_imd_file.ENERGYMIN
[15]:
ENERGYMIN
# NTEM NCYC DELE DX0 DXM NMIN FLIM
1 0 0.100000 0.010000 0.050000 2000 0.000000
END
SYSTEM Block¶
In the SYSTEM block you specify the number of solutes (NPM) and solvent (NSM) molecules. You only have one solute NPM = 1 and no solvent molecules NSM = 0 because you still did not add any solvent molecules to the configuration file and the peptide is still in vacuum. Otherwise you would have to tell MD++ how many solvent molecules you are using.
[16]:
help(emin_vac_imd_file.SYSTEM)
Help on SYSTEM in module pygromos.files.blocks.imd_blocks object:
class SYSTEM(_generic_imd_block)
| SYSTEM(NPM: int = 0, NSM: int = 0, content: List[str] = None)
|
| System Block
|
| The system block defines the number of solute molecules and solvent molecules
|
| Attributes
| ----------
| NPM: int
| Number of Solute Molecules
| NSM: int
| Number of Solvent Molecules
|
| Method resolution order:
| SYSTEM
| _generic_imd_block
| pygromos.files.blocks._general_blocks._generic_gromos_block
| builtins.object
|
| Methods defined here:
|
| __init__(self, NPM: int = 0, NSM: int = 0, content: List[str] = None)
| Initialize self. See help(type(self)) for accurate signature.
|
| ----------------------------------------------------------------------
| Data and other attributes defined here:
|
| __annotations__ = {'NPM': <class 'int'>, 'NSM': <class 'int'>, 'name':...
|
| name = 'SYSTEM'
|
| ----------------------------------------------------------------------
| Methods inherited from _generic_imd_block:
|
| block_to_string(self) -> str
|
| read_content_from_str(self, content: List[str])
|
| ----------------------------------------------------------------------
| Methods inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __copy__(self)
|
| __deepcopy__(self, memo)
|
| __eq__(self, _generic_gromos_block__o: object) -> bool
| Return self==value.
|
| __iter__(self)
|
| __repr__(self)
| Return repr(self).
|
| __str__(self)
| Return str(self).
|
| get_name(self)
|
| ----------------------------------------------------------------------
| Data descriptors inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
|
| ----------------------------------------------------------------------
| Data and other attributes inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __hash__ = None
[17]:
emin_vac_imd_file.SYSTEM
[17]:
SYSTEM
# NPM NSM
1 0
END
STEP Block¶
The step block takes core of how long and with
[18]:
help(emin_vac_imd_file.STEP)
Help on STEP in module pygromos.files.blocks.imd_blocks object:
class STEP(_generic_imd_block)
| STEP(NSTLIM: int = 0, T: float = 0, DT: float = 0, content: List[str] = None)
|
| Step Block
|
| This Block gives the number of simulation steps,
|
| Attributes
| -----------
| NSTLIM: int
| number of simulations Step till terminating.
| T: float
| Starting Time
| DT: float
| time step [fs]
|
| Method resolution order:
| STEP
| _generic_imd_block
| pygromos.files.blocks._general_blocks._generic_gromos_block
| builtins.object
|
| Methods defined here:
|
| __init__(self, NSTLIM: int = 0, T: float = 0, DT: float = 0, content: List[str] = None)
| Initialize self. See help(type(self)) for accurate signature.
|
| ----------------------------------------------------------------------
| Data and other attributes defined here:
|
| __annotations__ = {'DT': <class 'float'>, 'NSTLIM': <class 'int'>, 'T'...
|
| name = 'STEP'
|
| ----------------------------------------------------------------------
| Methods inherited from _generic_imd_block:
|
| block_to_string(self) -> str
|
| read_content_from_str(self, content: List[str])
|
| ----------------------------------------------------------------------
| Methods inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __copy__(self)
|
| __deepcopy__(self, memo)
|
| __eq__(self, _generic_gromos_block__o: object) -> bool
| Return self==value.
|
| __iter__(self)
|
| __repr__(self)
| Return repr(self).
|
| __str__(self)
| Return str(self).
|
| get_name(self)
|
| ----------------------------------------------------------------------
| Data descriptors inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
|
| ----------------------------------------------------------------------
| Data and other attributes inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __hash__ = None
[19]:
emin_vac_imd_file.STEP
[19]:
STEP
# NSTLIM T DT
3000 0.000000 0.002000
END
In the BOUNDCOND block you specify which periodic boundary conditions (PBC) you are going to use in the EM procedure. NTB = 0 defines a vacuum simulation: PBC are not applied. To indicate the truncated octahedron (t) PBC, NTB is set to -1, for rectangular (r) PBC NTB is 1, and for the triclinic (c) PBC NTB is 2. NDFMIN defines the number of degrees of freedom subtracted from the total number of degrees of freedom for the calculation of the temperature.
[20]:
help(emin_vac_imd_file.BOUNDCOND)
Help on BOUNDCOND in module pygromos.files.blocks.imd_blocks object:
class BOUNDCOND(_generic_imd_block)
| BOUNDCOND(NTB: int = 0, NDFMIN: int = 0, content: List[str] = None)
|
| Boundary Condition Block
|
| This block describes the boundary condition of the coordinate system.
|
| Attributes
| ----------
| NTB : int, optional
| Boundary conditions, by default 0
| -1 : truncated octahedral
| 0 : vacuum
| 1 : rectangular
| 2 : triclinic
| NDFMIN : int, optional
| number of degrees of freedom subtracted for temperature, by default 0
|
| Method resolution order:
| BOUNDCOND
| _generic_imd_block
| pygromos.files.blocks._general_blocks._generic_gromos_block
| builtins.object
|
| Methods defined here:
|
| __init__(self, NTB: int = 0, NDFMIN: int = 0, content: List[str] = None)
| Initialize self. See help(type(self)) for accurate signature.
|
| ----------------------------------------------------------------------
| Data and other attributes defined here:
|
| __annotations__ = {'NDFMIN': <class 'int'>, 'NTB': <class 'int'>, 'nam...
|
| name = 'BOUNDCOND'
|
| ----------------------------------------------------------------------
| Methods inherited from _generic_imd_block:
|
| block_to_string(self) -> str
|
| read_content_from_str(self, content: List[str])
|
| ----------------------------------------------------------------------
| Methods inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __copy__(self)
|
| __deepcopy__(self, memo)
|
| __eq__(self, _generic_gromos_block__o: object) -> bool
| Return self==value.
|
| __iter__(self)
|
| __repr__(self)
| Return repr(self).
|
| __str__(self)
| Return str(self).
|
| get_name(self)
|
| ----------------------------------------------------------------------
| Data descriptors inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
|
| ----------------------------------------------------------------------
| Data and other attributes inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __hash__ = None
[21]:
emin_vac_imd_file.BOUNDCOND
[21]:
BOUNDCOND
# NTB NDFMIN
0 0
END
With the PRINTOUT block you can specify how often (every NTPRth step) you are printing out the energies to the output file.
[22]:
help(emin_vac_imd_file.PRINTOUT)
Help on PRINTOUT in module pygromos.files.blocks.imd_blocks object:
class PRINTOUT(_generic_imd_block)
| PRINTOUT(NTPR: int = 0, NTPP: int = 0, content: List[str] = None)
|
| PRINTOUT block
|
| This Block manages the output frequency into the .omd/std-out file.
|
| Attributes
| ----------
| NTPR: int
| print out energies, etc. every NTPR steps
| NTPP: int
| =1 perform dihedral angle transition monitoring
|
| Method resolution order:
| PRINTOUT
| _generic_imd_block
| pygromos.files.blocks._general_blocks._generic_gromos_block
| builtins.object
|
| Methods defined here:
|
| __init__(self, NTPR: int = 0, NTPP: int = 0, content: List[str] = None)
| Initialize self. See help(type(self)) for accurate signature.
|
| ----------------------------------------------------------------------
| Data and other attributes defined here:
|
| __annotations__ = {'NTPP': <class 'int'>, 'NTPR': <class 'int'>, 'name...
|
| name = 'PRINTOUT'
|
| ----------------------------------------------------------------------
| Methods inherited from _generic_imd_block:
|
| block_to_string(self) -> str
|
| read_content_from_str(self, content: List[str])
|
| ----------------------------------------------------------------------
| Methods inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __copy__(self)
|
| __deepcopy__(self, memo)
|
| __eq__(self, _generic_gromos_block__o: object) -> bool
| Return self==value.
|
| __iter__(self)
|
| __repr__(self)
| Return repr(self).
|
| __str__(self)
| Return str(self).
|
| get_name(self)
|
| ----------------------------------------------------------------------
| Data descriptors inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
|
| ----------------------------------------------------------------------
| Data and other attributes inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __hash__ = None
[23]:
emin_vac_imd_file.PRINTOUT
[23]:
PRINTOUT
# NTPR NTPP
10 0
END
Bonds vibrate at high frequencies (hν ≫ kBT ). Therefore, these vibrations are of quantum-mechanical nature. So constraining the bond lengths is a better approximation than treating them as classical harmonic oscillators. Constraining all bond lengths of the solute and solvent (NTC=3) allows the use of a rather large time step of 2 fs. In this example the constraints are imposed by the SHAKE algorithm for both solute (NTCP=1) and solvent (NTCS=1) with a tolerance of 0.0001. See 4-90 for more information.
[24]:
help(emin_vac_imd_file.CONSTRAINT)
Help on CONSTRAINT in module pygromos.files.blocks.imd_blocks object:
class CONSTRAINT(_generic_imd_block)
| CONSTRAINT(NTC: int = 0, NTCP: int = 0, NTCP0: float = 0, NTCS: int = 0, NTCS0: float = 0, content: List[str] = None)
|
| CONSTRAINT block
| This block is controlling constraining the atoms during a simulation.
|
| Attributes
| ----------
| NTC: int
| NTCP: int
| NTCP0: int
| NTCS: int
| NTCS0: int
|
| Method resolution order:
| CONSTRAINT
| _generic_imd_block
| pygromos.files.blocks._general_blocks._generic_gromos_block
| builtins.object
|
| Methods defined here:
|
| __init__(self, NTC: int = 0, NTCP: int = 0, NTCP0: float = 0, NTCS: int = 0, NTCS0: float = 0, content: List[str] = None)
| Args:
| NTC:
| NTCP:
| NTCP0:
| NTCS:
| NTCS0:
|
| ----------------------------------------------------------------------
| Data and other attributes defined here:
|
| __annotations__ = {'NTC': <class 'int'>, 'NTCP': <class 'int'>, 'NTCP0...
|
| name = 'CONSTRAINT'
|
| ----------------------------------------------------------------------
| Methods inherited from _generic_imd_block:
|
| block_to_string(self) -> str
|
| read_content_from_str(self, content: List[str])
|
| ----------------------------------------------------------------------
| Methods inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __copy__(self)
|
| __deepcopy__(self, memo)
|
| __eq__(self, _generic_gromos_block__o: object) -> bool
| Return self==value.
|
| __iter__(self)
|
| __repr__(self)
| Return repr(self).
|
| __str__(self)
| Return str(self).
|
| get_name(self)
|
| ----------------------------------------------------------------------
| Data descriptors inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
|
| ----------------------------------------------------------------------
| Data and other attributes inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __hash__ = None
[25]:
emin_vac_imd_file.CONSTRAINT
[25]:
CONSTRAINT
# NTC
3
# NTCP NTCP0(1)
1 0.000100
# NTCS NTCS0(1)
1 0.000100
END
In the FORCE block you tell MD++ which terms it should use for the energy and force calculation. For bond angles, improper dihedrals, torsional dihedrals and the non-bonded interactions the standard terms of the GROMOS force field are switched on (1). Because we are using bond-length constraints and the SHAKE algorithm, we have to switch off (0) the bond-stretching terms for the bonds involving hydrogen atoms and not involving hydrogen atoms.. In the last line of this block, the energy groups are defined. In general, we define one or more energy groups for every molecule, and one comprising all the solvent molecules. The first integer is the number of energy groups we want to use (in the present case we only have one energy group). The following numbers are the atom sequence numbers of the last atom of each energy group. By defining these energy groups we 7-9 tell MD++ to sum up the energies between the atoms within these groups and calculate the inter-group energies, which can be very useful.
[26]:
emin_vac_imd_file.FORCE
[26]:
FORCE
# BONDS ANGLES IMPROPER DIHEDRAL ELECTROSTATIC VDW
0 1 1 1 1 1
# NEGR NRE
1 71
END
In the PAIRLIST block you specify which algorithm you will use for the pairlist generation. The cut-off used in the short-range pairlist construction is given by RCUTP and for GROMOS it is usually 0.8 nm. The cut-off used in the long-range interactions is given by RCUTL and for GROMOS it is usually 1.4 nm. The pairlist is generated every 5th (NSNB) step. TYPE specifies the type of the cut-off, whether it is based on the distance between charge-groups (0) or on the distance between atoms (1).
Warning: Think very carefully about the definition of energy groups before running the simulation. Energies of energy groups can not be calculated from the trajectories in an efficient way. So, changing an energy-group definition will result in rerunning the simulation. ***
[27]:
help(emin_vac_imd_file.PAIRLIST)
Help on PAIRLIST in module pygromos.files.blocks.imd_blocks object:
class PAIRLIST(_generic_imd_block)
| PAIRLIST(ALGORITHM: int = 0, NSNB: int = 0, RCUTP: float = 0, RCUTL: float = 0, SIZE: Union[str, float] = 0, TYPE: Union[str, bool] = False, content: List[str] = None)
|
| PAIRLIST Block
|
| This block is controlling the pairlist control.
|
| Attributes
| ----------
| ALGORITHM: int
| standard(0) (gromos96 like pairlist)
| grid(1) (md++ grid pairlist)
| grid_cell(2) (creates a mask)
| NSNB: int
| frequency (number of steps) a pairlist is constructed
| RCUTPL: float
| short-range cut-off in twin-range
| RCUTL: float
| intermediate-range cut-off in twin-range
| SIZE: str, float
| grid cell size (or auto = 0.5 * RCUTP)
| TYPE: str, bool
| chargegoup(0) (chargegroup based cutoff)
| atomic(1) (atom based cutoff)
|
| Method resolution order:
| PAIRLIST
| _generic_imd_block
| pygromos.files.blocks._general_blocks._generic_gromos_block
| builtins.object
|
| Methods defined here:
|
| __init__(self, ALGORITHM: int = 0, NSNB: int = 0, RCUTP: float = 0, RCUTL: float = 0, SIZE: Union[str, float] = 0, TYPE: Union[str, bool] = False, content: List[str] = None)
| Args:
| ALGORITHM:
| NSNB:
| RCUTP:
| RCUTL:
| SIZE:
| TYPE:
|
| ----------------------------------------------------------------------
| Data and other attributes defined here:
|
| __annotations__ = {'ALGORITHM': <class 'int'>, 'NSNB': <class 'int'>, ...
|
| name = 'PAIRLIST'
|
| ----------------------------------------------------------------------
| Methods inherited from _generic_imd_block:
|
| block_to_string(self) -> str
|
| read_content_from_str(self, content: List[str])
|
| ----------------------------------------------------------------------
| Methods inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __copy__(self)
|
| __deepcopy__(self, memo)
|
| __eq__(self, _generic_gromos_block__o: object) -> bool
| Return self==value.
|
| __iter__(self)
|
| __repr__(self)
| Return repr(self).
|
| __str__(self)
| Return str(self).
|
| get_name(self)
|
| ----------------------------------------------------------------------
| Data descriptors inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
|
| ----------------------------------------------------------------------
| Data and other attributes inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __hash__ = None
[28]:
emin_vac_imd_file.PAIRLIST
[28]:
PAIRLIST
# ALGORITHM NSNB RCUTP RCUTL SIZE TYPE
0 5 0.800000 1.400000 0.4 0
END
In the NONBONDED block you specify using NLRELE which method for the evaluation of long-range electrostatic interactions is used. Since you will use the reaction-field method, the value of NLRELE should be equal to 1. The long-range electrostatic interactions are truncated beyond a certain cutoff (RCUTL in the PAIRLIST block). Beyond the reaction-field cut-off radius (RCRF) the electrostatic interactions are replaced by a static reaction field with a dielectric permittivity of EPSRF. RCRF and RCUTL should be identical. Because we are doing the energy minimisation in vacuo EPSRF is set to 1. With NSLFEXCL equal to 1, you include the contributions of excluded atoms to the electrostatic energy. The ionic strength of the continuum is set to 0 (APPAK). All other switches are not used for the reaction-field method. See 4-98 for more information.
[29]:
help(emin_vac_imd_file.NONBONDED)
Help on NONBONDED in module pygromos.files.blocks.imd_blocks object:
class NONBONDED(_generic_imd_block)
| NONBONDED(NLRELE: int = 0, APPAK: float = 0, RCRF: float = 0, EPSRF: float = 0, NSLFEXCL: bool = False, NSHAPE: int = 0, ASHAPE: float = 0, NA2CLC: int = 0, TOLA2: str = 0, EPSLS: float = 0, NKX: int = 0, NKY: int = 0, NKZ: int = 0, KCUT: float = 0, NGX: int = 0, NGY: int = 0, NGZ: int = 0, NASORD: int = 0, NFDORD: int = 0, NALIAS: int = 0, NSPORD: int = 0, NQEVAL: int = 0, FACCUR: float = 0, NRDGRD: bool = False, NWRGRD: bool = False, NLRLJ: bool = False, SLVDNS: float = 0, content: List[str] = None)
|
| NONBONDED block
|
| This block is controlling the Nonbonded term evaluation
|
| Attributes
| ----------
| NLRELE: int
| 1-3 method to handle electrostatic interactions
| -1 : reaction-field (LSERF compatibility mode)
| 0 : no electrostatic interactions
| 1 : reaction-field
| 2 : Ewald method
| 3 : P3M method
| APPAK: float
| >= 0.0 reaction-field inverse Debye screening length
| RCRF: float
| >= 0.0 reaction-field radius
| EPSRF: float
| = 0.0 || > 1.0 reaction-field permittivity
| NSLFEXCL: bool
| contribution of excluded atoms to reaction field false=off/true=on
| NSHAPE: float
| -1..10 lattice sum charge-shaping function
| -1 : gaussian
| 0..10 : polynomial
| ASHAPE: float
| > 0.0 width of the lattice sum charge-shaping function
| NA2CLC: int
| 0..4 controls evaluation of lattice sum A2 term
| 0 : A2 = A2~ = 0
| 1 : A2~ exact, A2 = A2~
| 2 : A2 numerical, A2~ = A2
| 3 : A2~ exact from Ewald or from mesh and atom coords, A2 numerical
| 4 : A2~ averaged from mesh only, A2 numerical
| TOLA2: float
| > 0.0 tolerance for numerical A2 evaluation
| EPSLS: float
| = 0.0 || > 1.0 lattice sum permittivity (0.0 = tinfoil)
| NKX, NKY, NKZ: float
| > 0 maximum absolute Ewald k-vector components
| KCUT: float
| > 0.0 Ewald k-space cutoff
| NGX, NGY, NGZ: float
| > 0 P3M number of grid points
| NASORD: int
| 1..5 order of mesh charge assignment function
| NFDORD: int
| 0..5 order of the mesh finite difference operator
| 0 : ik - differentiation
| 1..5 : finite differentiation
| NALIAS: float
| > 0 number of mesh alias vectors considered
| NSPORD: float
| order of SPME B-spline functions (not available)
| NQEVAL: float
| >= 0 controls accuracy reevaluation
| 0 : do not reevaluate
| > 0 : evaluate every NQEVAL steps
| FACCUR: float
| > 0.0 rms force error threshold to recompute influence function
| NRDGRD: bool
| 0,1 read influence function
| 0 : calculate influence function at simulation start up
| 1 : read influence function from file (not yet implemented)
| NWRGRD: bool
| 0,1 write influence function
| 0 : do not write
| 1 : write at the end of the simulation (not yet implemented)
| NLRLJ: bool
| 0,1 controls long-range Lennard-Jones corrections
| 0 : no corrections
| 1 : do corrections (not yet implemented)
| SLVDNS: float
| > 0.0 average solvent density for long-range LJ correction (ignored)
|
| Method resolution order:
| NONBONDED
| _generic_imd_block
| pygromos.files.blocks._general_blocks._generic_gromos_block
| builtins.object
|
| Methods defined here:
|
| __init__(self, NLRELE: int = 0, APPAK: float = 0, RCRF: float = 0, EPSRF: float = 0, NSLFEXCL: bool = False, NSHAPE: int = 0, ASHAPE: float = 0, NA2CLC: int = 0, TOLA2: str = 0, EPSLS: float = 0, NKX: int = 0, NKY: int = 0, NKZ: int = 0, KCUT: float = 0, NGX: int = 0, NGY: int = 0, NGZ: int = 0, NASORD: int = 0, NFDORD: int = 0, NALIAS: int = 0, NSPORD: int = 0, NQEVAL: int = 0, FACCUR: float = 0, NRDGRD: bool = False, NWRGRD: bool = False, NLRLJ: bool = False, SLVDNS: float = 0, content: List[str] = None)
| Initialize self. See help(type(self)) for accurate signature.
|
| ----------------------------------------------------------------------
| Data and other attributes defined here:
|
| __annotations__ = {'APPAK': <class 'float'>, 'ASHAPE': <class 'float'>...
|
| name = 'NONBONDED'
|
| ----------------------------------------------------------------------
| Methods inherited from _generic_imd_block:
|
| block_to_string(self) -> str
|
| read_content_from_str(self, content: List[str])
|
| ----------------------------------------------------------------------
| Methods inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __copy__(self)
|
| __deepcopy__(self, memo)
|
| __eq__(self, _generic_gromos_block__o: object) -> bool
| Return self==value.
|
| __iter__(self)
|
| __repr__(self)
| Return repr(self).
|
| __str__(self)
| Return str(self).
|
| get_name(self)
|
| ----------------------------------------------------------------------
| Data descriptors inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
|
| ----------------------------------------------------------------------
| Data and other attributes inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __hash__ = None
[30]:
emin_vac_imd_file.NONBONDED
[30]:
NONBONDED
# NLRELE
1
# APPAK RCRF EPSRF NSLFEXCL
0.000000 1.400000 61.000000 1
# NSHAPE ASHAPE NA2CLC TOLA2 EPSLS
-1 1.400000 2 1e-10 0.000000
# NKX NKY NKZ KCUT
10 10 10 100.000000
# NGX NGY NGZ NASORD NFDORD NALIAS NSPORD
32 32 32 3 2 3 4
# NQEVAL FACCUR NRDGRD NWRGRD
100000 1.600000 0 0
# NLRLJ SLVDNS
0 33.300000
END
Perform Emin¶
In order to run the MD++ program, a shell script needs to be prepared. Open the shell script em peptide. run and adapt the paths and the names of the files according to your system.
[31]:
from pygromos.simulations.modules.preset_simulation_modules import emin
out_prefix = "emin_vacuum"
## Preparing emin gromos system
in_emin_system = build_system.copy()
in_emin_system.work_folder = project_dir
in_emin_system._gromosXX_bin_dir = gromosXX_bin
in_emin_system.imd = emin_vac_imd_file # This step is not necessary, as we did this before
from pygromos.files.blocks.imd_blocks import WRITETRAJ
in_emin_system.imd.add_block(block=WRITETRAJ(NTWE=25, NTWX=25))
in_emin_system.prepare_for_simulation()
in_emin_system
[31]:
GROMOS SYSTEM: peptideH
################################################################################
WORKDIR: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System
LAST CHECKPOINT: None
GromosXX_bin: None
GromosPP_bin: None
FILES:
imd: /home/mlehner/PyGromosTools/pygromos/data/simulation_parameters_templates/vac_emin.imd
top: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/peptideH.top
cnf: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/peptideH.cnf
FUTURE PROMISE: False
SYSTEM:
PROTEIN: protein nresidues: 5 natoms: 71
LIGANDS: [] resID: [] natoms: 0
Non-LIGANDS: [] nmolecules: 0 natoms: 0
SOLVENT: [] nmolecules: 0 natoms: 0
[32]:
out_emin_system = emin(in_gromos_system=in_emin_system,
step_name="b_"+out_prefix)
################################################################################
b_emin_vacuum
################################################################################
Script: /home/mlehner/PyGromosTools/pygromos/simulations/hpc_queuing/job_scheduling/schedulers/simulation_scheduler.py
################################################################################
Simulation Setup:
################################################################################
steps_per_run: 3000
equis: 0
simulation runs: 1
################################################################################
submit final analysis part
/home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/b_emin_vacuum/ana_out.log
/home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/b_emin_vacuum/job_analysis.py
ANA jobID: 0
Analysis¶
Once the energy minimisation is finished, the file with the minimized coordinates, peptide min.cnf, and the general output file, em peptide.omd, that reports the progress of the minimisation, will be written out. Have a look at both files and check if the minimisation has finished successfully.
[33]:
out_emin_system.cnf.view
This is nice, but how do we actually get to this structure! In the next cell the development of the total Potential energy over the energyminimization steps is shown. As you can see, the Potential energy of the whole system is decreasing, which indicates a relaxatio of the peptide structure and an optimization of the different forcefield terms. Ergo finding a minimal total potential system energy as possible is desirable in energyminimizations.
[34]:
out_emin_system.tre.get_totals().totene.plot(xlabel="steps", ylabel="$V_{tot}~[kJ/mol]$")
[34]:
<AxesSubplot:xlabel='steps', ylabel='$V_{tot}~[kJ/mol]$'>
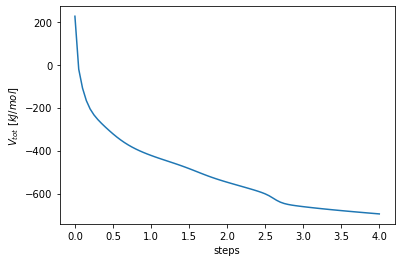
The effect seend in the total potential energy can also be observed of course in the peptide molecue coordinats.
[35]:
view = out_emin_system.trc.view
view
/home/mlehner/anaconda3/envs/pygro2/lib/python3.9/site-packages/mdtraj/core/trajectory.py:438: UserWarning: top= kwargs ignored since this file parser does not support it
warnings.warn('top= kwargs ignored since this file parser does not support it')
Finally we wan to store our energy minimization results as a python pickle obj.
[36]:
out_emin_system_path = out_emin_system.save(out_emin_system.work_folder+"/emin_vac_result.obj")
[37]:
#if you want to load the file:
#out_emin_system = Gromos_System.load(out_emin_system.work_folder+"/emin_vac_result.obj")
Solvatistation and Solvent Energy Minimization¶
Now you can put the energy minimized penta-peptide in a box of solvent to simulate an aequos environment. This can be done with the sim_box function of the gromosSyste, which can solvate the solute in a pre-equilibrated box of solvent molecules. As input for the function sim_box you have to specify the following input arguments: * periodic_boundary_condition: the resulting box shape under the argument (r - rectangular, t - truncated octahedron, c - triclinic) * in_solvent_cnf_file_path: the coordinate file of the pre-equilibrated box of solvent molecules under the argument (we are using a template \(H_2O\) of the SPC model provided in pygromos.data.solvent_coordinates) * minwall: minimum solute-to-wall distance in \(nm\) * threshold: the minimum solute-to-solvent distance ins \(nm\) * rotate: If you are using a rectangular box (@pbc r) it is recommended to use an additional argument. With this additional argument the solute can be rotated (before solvating) such that the largest distance between any two solute atoms is directed along the z-axis, and the largest atom-atom distance in the xy-plane lies in the y-direction. An input file sim box peptide.arg is already prepared.
[38]:
help(out_emin_system.sim_box)
Help on function sim_box in module pygromos.gromos.gromosPP:
sim_box(in_solvent_cnf_file_path: str, periodic_boundary_condition: str = 'r', gathering_method: str = None, minwall: float = 0.8, threshold: float = None, rotate: str = None, boxsize: bool = False, _binary_name: str = 'sim_box', verbose: bool = False) -> str
When simulating a molecule in solution or in a crystal containing solvent
molecules, the atomic coordinates of the solvent molecules are to be
generated, if they are not available from experiment. Program sim_box can
solvate a solute in a pre-equilibrated box of solvent molecules. The file
specifying the solvent configuration should contain a BOX block with the
dimensions corresponding to the pre-equilibrated density. The solvent
topology is read from the solvent block in the specified topology.
Parameters
----------
in_top_path : str
the path to the input topology file (.top)
in_cnf_path : str
the path to the input coordinate file (.cnf), which shall be solvated
in_solvent_cnf_file_path : str
the path to the input coordinate file of the solvent (.cnf), that shall be used to solvate (checkout pygromos.data.solvent_coordinates for templates)
out_cnf_path : str, optional
the path to the resulting coordinate (.cnf) file, by default ""
periodic_boundary_condition : str, optional
describes the boundary condition of the given system in the cnf. (r - rectangle, v - vacuum, ), by default "r"
gathering_method : str, optional
the gathering method to be used, by default None
minwall : float, optional
minimum solute to wall distance, by default 0.8
threshold : float, optional
minimum solvent-solute distance, by default None -> 0.23 nm
rotate : str, optional
rotate solute: biggest axis along z, second along y, by default None
boxsize : bool, optional
use boxsize specified in solute coordinate file, by default False
_binary_name : str, optional
name of the binary, by default "sim_box"
verbose : bool, optional
stay a while and listen!, by default False
Returns
-------
str
return the path to the resulting cnf path.
[39]:
vars(out_emin_system.trc).keys()
[39]:
dict_keys(['_future_file', '_topology', '_xyz', '_rmsd_traces', '_unitcell_lengths', '_unitcell_angles', '_time_default_to_arange', '_time', '_step', 'TITLE', 'path', '_view'])
build box system¶
To put the solvent box around the penta-peptide use following commands:
[40]:
from pygromos.data.solvent_coordinates import spc
#setup a fresh gromos System:
box_system = out_emin_system.copy()
#box_system = gromos_system.Gromos_System.load(out_emin_system_path) #if you do this step after a break, you could also decide to load the serialized obj from before.
box_system.imd = None
box_system.name = "solvate_box"
box_system.work_folder = bash.make_folder(project_dir+"/c_"+box_system.name)
## set box and solvate the system
box_system.cnf.add_empty_box()
box_system.sim_box(in_solvent_cnf_file_path=spc,
periodic_boundary_condition="r",
minwall=0.8,
threshold=0.23,
rotate=False)
#box_system.cnf
box_system.rebase_files() #write out the files (optional) - so you can check them in the folder
box_system.cnf.view #show the results
/home/mlehner/PyGromosTools/pygromos/files/gromos_system/gromos_system.py:1123: UserWarning: did not find the required imd found
warnings.warn("did not find the required " + file_name + " found")
Add Ions¶
After solvating the system, two \(Cl-\) ions shall be added to the box as counter charges for the two positive peptide residues (Arginine and Lysine).
To add the ions, we need to know the name of the building block in Gromos54A7. We can checkout the names of the Atomtype names in the topology of our system:
[41]:
print([ a for a in box_system.top.ATOMTYPENAME])
[['54'], ['O'], ['OM'], ['OA'], ['OE'], ['OW'], ['N'], ['NT'], ['NL'], ['NR'], ['NZ'], ['NE'], ['C'], ['CH0'], ['CH1'], ['CH2'], ['CH3'], ['CH4'], ['CH2r'], ['CR1'], ['HC'], ['H'], ['DUM'], ['S'], ['CU1+'], ['CU2+'], ['FE'], ['ZN2+'], ['MG2+'], ['CA2+'], ['P,SI'], ['AR'], ['F'], ['CL'], ['BR'], ['CMet'], ['OMet'], ['NA+'], ['CL-'], ['CChl'], ['CLChl'], ['HChl'], ['SDmso'], ['CDmso'], ['ODmso'], ['CCl4'], ['CLCl4'], ['FTFE'], ['CTFE'], ['CHTFE'], ['OTFE'], ['CUrea'], ['OUrea'], ['NUrea'], ['CH3p']]
After finding the atomtype name (hint: ‘CL-’), we next can use it to add two of these ions to our system with the ion function.
[42]:
help(box_system.ion)
Help on function ion in module pygromos.gromos.gromosPP:
ion(in_building_block_lib_path: str, in_parameter_lib_path: str, periodic_boundary_condition: str = 'v', negative: list = None, positive: list = None, potential: float = 0.8, mindist: float = 0.8, random_seed: int = None, exclude: str = None, _binary_name: str = 'ion', verbose: bool = False) -> str
When simulating a charged solute in solution, one may wish to include
counter-ions in the molecular system in order to obtain a neutral system, or
a system with a specific ionic strength. The program ion can replace solvent
molecules by atomic ions by placing the
ion at the position of the first atom of a solvent molecule. Substitution of
solvent molecules by positive or negative ions can be performed by selecting
the solvent positions with the lowest or highest Coulomb potential, respectively,
or by random selection. In order to prevent two ions being placed too
close together, a sphere around each inserted ion can be specified from which
no solvent molecules will be substituted by additional ions. In addition, the user can
specify specific water molecules that should not be considered for
replacement.
Parameters
----------
in_top_path : str
the path to the input topology file (.top)
in_cnf_path : str
the path to the input coordinate file (.cnf), to which the ions shall be added
out_cnf_path : str
the path to the resulting coordinate (.cnf) file
periodic_boundary_condition : str, optional
describes the boundary condition of the given system in the cnf. (r - rectangle, v - vacuum, ). a gathering method can be optionally added with a whitespace seperation., by default "v"
negative : list, optional
the first element of the list is the number of ions and the second element of the list is the type of ion, optionally a third element can be passed giving the residue name, by default None
positive : list, optional
the first element of the list is the number of ions and the second element of the list is the type of ion, optionally a third element can be passed giving the residue name, by default None
potential : float, optional
cutoff for potential calculation[nm], by default 0.8
mindist : float, optional
minimum distance between ions[nm], by default 0.8
random_seed : int, optional
provide the used random seed, by default None
exclude : str, optional
if you want to exclude solvent molecules, define a gromos selection here, by default None
_binary_name : str, optional
the program name, by default "ion"
verbose : bool, optional
stay a while and listen, by default False
Returns
-------
str
returns the resulting cnf-file path
[43]:
from pygromos.data.solvent_coordinates import spc
## Build directory and setup a fresh Gromos System
ion_system = box_system.copy()
ion_system.name = "ion"
ion_system.work_folder = bash.make_folder(project_dir+"/d_"+ion_system.name)
#Add the ions to the System
ion_system.ion(negative=[2, "CL-"])
ion_system.rebase_files() #write out the files, so you can check them (optional)
ion_system.cnf.view #show the results.
Energy Minimization BOX¶
In order to relax the unfavorable atom-atom contacts between the solute and the solvent, energy minimization of the solvent should be performed while keeping the solute positionally restrained (i.e. connecting the atom to a reference position by a spring). In order to do that two additional files, in which the positionally restrained atoms and the reference coordinates are specified, have to be generated from the coordinate file in our gromos system. Afterwards, we are going to run the second energy minimization
build posistion restraints¶
To apply position restraints to our simulation for the peptide, we first need to generate a selection of residues, that shall not be modified by the energyminimization. This translates to the residues of the peptide. After this the function generate_posres can be used to generate two files. * position restraints - posres(.por) This file defines the selection of atoms, that shall be restrained during the simulation. * reference position - refpos (.rpf) This files defines a reference position for the system. Gromos will restrain the selected atoms in the posres file to the reference position file. So if the atom position during the simulation deviates from the refpos file, the program will restrain the atom, such it moves towards the reference position.
[44]:
from pygromos.simulations.modules.preset_simulation_modules import emin
from pygromos.data.simulation_parameters_templates import template_emin
# Preparing emin gromos system
in_eminBox_system = ion_system.copy()
in_eminBox_system.name = "emin_solvBox"
in_eminBox_system.work_folder = project_dir
# Build position restraints
## Build selection for residues
restrain_res = [k for k in in_eminBox_system.cnf.residues if(not k in ("CL-", "SOLV"))]
print("Selection of residues: ", restrain_res)
## Build the restrain files
in_eminBox_system.generate_posres(residues=restrain_res)
# Check simulation params
in_eminBox_system.imd = template_emin #Here we use template simulation parameters, The blocks are the same as above in the vacuum case with slight deviations.
in_eminBox_system.imd.INITIALISE.NTISHI = 1
in_eminBox_system.prepare_for_simulation()
in_eminBox_system
Selection of residues: ['VAL', 'TYR', 'ARG', 'LYSH', 'GLN']
[44]:
GROMOS SYSTEM: emin_solvBox
################################################################################
WORKDIR: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System
LAST CHECKPOINT: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/emin_vac_result.obj
GromosXX_bin: None
GromosPP_bin: None
FILES:
imd: /home/mlehner/PyGromosTools/pygromos/data/simulation_parameters_templates/emin.imd
top: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/emin_solvBox.top
cnf: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/emin_solvBox.cnf
posres: None
refpos: None
FUTURE PROMISE: False
SYSTEM:
PROTEIN: protein nresidues: 5 natoms: 71
LIGANDS: ['CL6', 'CL7'] resID: [6, 7] natoms: 2
Non-LIGANDS: [] nmolecules: 0 natoms: 0
SOLVENT: SOLV nmolecules: 930 natoms: 2790
In the ouptut of the in_eminBox_system two new files appeared the posres and refpos file:
[45]:
print(in_eminBox_system.posres)
TITLE
ion has replaced 2 solvent molecules in /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/d_ion/ion.cnf by
2 negative ions (CL-)
Solvent molecules were selected according to the highest / lowest
electrostatic potential
>>> Generated with PyGromosTools (riniker group) <<<
END
POSRESSPEC
#
1 VAL H1 1 -0.466039509 0.107068229 -0.151069220
1 VAL H2 2 -0.401465786 0.026940681 -0.024813902
1 VAL N 3 -0.432414924 0.018324392 -0.119519889
1 VAL H3 4 -0.505798596 -0.049378313 -0.125221767
1 VAL CA 5 -0.318752364 -0.024224333 -0.202467114
1 VAL CB 6 -0.373465010 -0.031635286 -0.345171611
1 VAL CG1 7 -0.375327181 0.100384420 -0.422508155
1 VAL CG2 8 -0.312775396 -0.149655615 -0.421315267
1 VAL C 9 -0.202187437 0.069532034 -0.170330753
1 VAL O 10 -0.203245639 0.121386106 -0.058800328
2 TYR N 11 -0.102128907 0.084161731 -0.256737514
2 TYR H 12 -0.098080944 0.029885412 -0.340636582
2 TYR CA 13 0.018655880 0.167364265 -0.246683687
2 TYR CB 14 -0.002756278 0.314387014 -0.283236038
2 TYR CG 15 -0.048773935 0.406717195 -0.170250028
2 TYR CD1 16 -0.182678567 0.413240325 -0.133567598
2 TYR HD1 17 -0.260065781 0.384896607 -0.204888104
2 TYR CD2 18 0.048327903 0.468975895 -0.092686587
2 TYR HD2 19 0.151325797 0.473772485 -0.128020125
2 TYR CE1 20 -0.217487257 0.473004714 -0.013000399
2 TYR HE1 21 -0.322783097 0.484771339 0.012564216
2 TYR CE2 22 0.013175251 0.530464601 0.026900147
2 TYR HE2 23 0.089050177 0.587521539 0.080458541
2 TYR CZ 24 -0.119337180 0.527807813 0.068756454
2 TYR OH 25 -0.145507035 0.540882621 0.201568903
2 TYR HH 26 -0.081715914 0.607922948 0.239449083
2 TYR C 27 0.115805998 0.136914297 -0.132474868
2 TYR O 28 0.211640835 0.063818891 -0.157056503
3 ARG N 29 0.083151951 0.165250087 -0.006686980
3 ARG H 30 0.001388625 0.218814191 0.014470360
3 ARG CA 31 0.172035422 0.131181336 0.105343321
3 ARG CB 32 0.160963320 0.232822082 0.219172617
3 ARG CG 33 0.031760480 0.225513789 0.300801725
3 ARG CD 34 0.065987501 0.269750448 0.443226804
3 ARG NE 35 0.003769017 0.179854770 0.541485965
3 ARG HE 36 -0.091497332 0.201673696 0.562658053
3 ARG CZ 37 0.062141586 0.075048175 0.601186018
3 ARG NH1 38 0.184794140 0.032263967 0.568343747
3 ARG HH11 39 0.228843408 -0.042435179 0.618121597
3 ARG HH12 40 0.228807480 0.065885805 0.485091775
3 ARG NH2 41 -0.001260231 0.013058464 0.701645134
3 ARG HH21 42 0.041195663 -0.065563290 0.746532886
3 ARG HH22 43 -0.089541851 0.046195549 0.734918729
3 ARG C 44 0.173254971 -0.013870953 0.154028761
3 ARG O 45 0.185208597 -0.042309231 0.273103508
4 LYSH N 46 0.162807050 -0.103932982 0.056721659
4 LYSH H 47 0.182603907 -0.070849428 -0.035551490
4 LYSH CA 48 0.156904142 -0.250588582 0.065120742
4 LYSH CB 49 0.048571437 -0.300415450 0.160991537
4 LYSH CG 50 0.085934868 -0.439794674 0.211848260
4 LYSH CD 51 -0.037748891 -0.523515797 0.245045132
4 LYSH CE 52 0.005417510 -0.661279435 0.295709174
4 LYSH NZ 53 -0.108328480 -0.754275429 0.300034383
4 LYSH HZ1 54 -0.135609930 -0.776899716 0.206528187
4 LYSH HZ2 55 -0.185192371 -0.711020046 0.347158498
4 LYSH HZ3 56 -0.081715914 -0.837556030 0.348574218
4 LYSH C 57 0.136972566 -0.313251518 -0.073044194
4 LYSH O 58 0.031106181 -0.367340392 -0.104625036
5 GLN N 59 0.236073002 -0.287496691 -0.157925246
5 GLN H 60 0.316859105 -0.234277179 -0.132579675
5 GLN CA 61 0.263132888 -0.332739421 -0.295157267
5 GLN CB 62 0.160944769 -0.282204560 -0.397220388
5 GLN CG 63 0.167762938 -0.132321877 -0.427113398
5 GLN CD 64 0.066161499 -0.088257923 -0.532680987
5 GLN OE1 65 -0.042879991 -0.039560951 -0.503277398
5 GLN NE2 66 0.105103739 -0.093861261 -0.659724117
5 GLN HE21 67 0.041195663 -0.065563290 -0.731234275
5 GLN HE22 68 0.197931140 -0.125751231 -0.678828780
5 GLN C 69 0.402663055 -0.277858242 -0.325644310
5 GLN O1 70 0.448562786 -0.295773090 -0.440523695
5 GLN O2 71 0.458569483 -0.212000520 -0.235286858
END
[46]:
in_eminBox_system.refpos
[46]:
TITLE
ion has replaced 2 solvent molecules in /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/d_ion/ion.cnf by
2 negative ions (CL-)
Solvent molecules were selected according to the highest / lowest
electrostatic potential
>>> Generated with PyGromosTools (riniker group) <<<
END
REFPOSITION
#
1 VAL H1 1 -0.466039509 0.107068229 -0.151069220
1 VAL H2 2 -0.401465786 0.026940681 -0.024813902
1 VAL N 3 -0.432414924 0.018324392 -0.119519889
1 VAL H3 4 -0.505798596 -0.049378313 -0.125221767
1 VAL CA 5 -0.318752364 -0.024224333 -0.202467114
1 VAL CB 6 -0.373465010 -0.031635286 -0.345171611
1 VAL CG1 7 -0.375327181 0.100384420 -0.422508155
1 VAL CG2 8 -0.312775396 -0.149655615 -0.421315267
1 VAL C 9 -0.202187437 0.069532034 -0.170330753
1 VAL O 10 -0.203245639 0.121386106 -0.058800328
2 TYR N 11 -0.102128907 0.084161731 -0.256737514
2 TYR H 12 -0.098080944 0.029885412 -0.340636582
2 TYR CA 13 0.018655880 0.167364265 -0.246683687
2 TYR CB 14 -0.002756278 0.314387014 -0.283236038
2 TYR CG 15 -0.048773935 0.406717195 -0.170250028
2 TYR CD1 16 -0.182678567 0.413240325 -0.133567598
2 TYR HD1 17 -0.260065781 0.384896607 -0.204888104
2 TYR CD2 18 0.048327903 0.468975895 -0.092686587
2 TYR HD2 19 0.151325797 0.473772485 -0.128020125
2 TYR CE1 20 -0.217487257 0.473004714 -0.013000399
2 TYR HE1 21 -0.322783097 0.484771339 0.012564216
2 TYR CE2 22 0.013175251 0.530464601 0.026900147
2 TYR HE2 23 0.089050177 0.587521539 0.080458541
2 TYR CZ 24 -0.119337180 0.527807813 0.068756454
2 TYR OH 25 -0.145507035 0.540882621 0.201568903
2 TYR HH 26 -0.081715914 0.607922948 0.239449083
2 TYR C 27 0.115805998 0.136914297 -0.132474868
2 TYR O 28 0.211640835 0.063818891 -0.157056503
3 ARG N 29 0.083151951 0.165250087 -0.006686980
3 ARG H 30 0.001388625 0.218814191 0.014470360
3 ARG CA 31 0.172035422 0.131181336 0.105343321
3 ARG CB 32 0.160963320 0.232822082 0.219172617
3 ARG CG 33 0.031760480 0.225513789 0.300801725
3 ARG CD 34 0.065987501 0.269750448 0.443226804
3 ARG NE 35 0.003769017 0.179854770 0.541485965
3 ARG HE 36 -0.091497332 0.201673696 0.562658053
3 ARG CZ 37 0.062141586 0.075048175 0.601186018
3 ARG NH1 38 0.184794140 0.032263967 0.568343747
3 ARG HH11 39 0.228843408 -0.042435179 0.618121597
3 ARG HH12 40 0.228807480 0.065885805 0.485091775
3 ARG NH2 41 -0.001260231 0.013058464 0.701645134
3 ARG HH21 42 0.041195663 -0.065563290 0.746532886
3 ARG HH22 43 -0.089541851 0.046195549 0.734918729
3 ARG C 44 0.173254971 -0.013870953 0.154028761
3 ARG O 45 0.185208597 -0.042309231 0.273103508
4 LYSH N 46 0.162807050 -0.103932982 0.056721659
4 LYSH H 47 0.182603907 -0.070849428 -0.035551490
4 LYSH CA 48 0.156904142 -0.250588582 0.065120742
4 LYSH CB 49 0.048571437 -0.300415450 0.160991537
4 LYSH CG 50 0.085934868 -0.439794674 0.211848260
4 LYSH CD 51 -0.037748891 -0.523515797 0.245045132
4 LYSH CE 52 0.005417510 -0.661279435 0.295709174
4 LYSH NZ 53 -0.108328480 -0.754275429 0.300034383
4 LYSH HZ1 54 -0.135609930 -0.776899716 0.206528187
4 LYSH HZ2 55 -0.185192371 -0.711020046 0.347158498
4 LYSH HZ3 56 -0.081715914 -0.837556030 0.348574218
4 LYSH C 57 0.136972566 -0.313251518 -0.073044194
4 LYSH O 58 0.031106181 -0.367340392 -0.104625036
5 GLN N 59 0.236073002 -0.287496691 -0.157925246
5 GLN H 60 0.316859105 -0.234277179 -0.132579675
5 GLN CA 61 0.263132888 -0.332739421 -0.295157267
5 GLN CB 62 0.160944769 -0.282204560 -0.397220388
5 GLN CG 63 0.167762938 -0.132321877 -0.427113398
5 GLN CD 64 0.066161499 -0.088257923 -0.532680987
5 GLN OE1 65 -0.042879991 -0.039560951 -0.503277398
5 GLN NE2 66 0.105103739 -0.093861261 -0.659724117
5 GLN HE21 67 0.041195663 -0.065563290 -0.731234275
5 GLN HE22 68 0.197931140 -0.125751231 -0.678828780
5 GLN C 69 0.402663055 -0.277858242 -0.325644310
5 GLN O1 70 0.448562786 -0.295773090 -0.440523695
5 GLN O2 71 0.458569483 -0.212000520 -0.235286858
6 CL- CL- 72 -0.498584400 -0.238913847 -0.020900428
7 CL- CL- 73 -0.302768665 0.293656105 0.699788447
1 SOLV OW 74 1.326391995 -1.191127515 -0.190725428
1 SOLV HW1 75 1.246610283 -1.250363472 -0.179433959
1 SOLV HW2 76 1.342544714 -1.175201155 -0.288126253
2 SOLV OW 77 -0.804324664 0.483084746 1.116278463
2 SOLV HW1 78 -0.795884454 0.575557397 1.079180521
2 SOLV HW2 79 -0.743545314 0.421412459 1.066267106
3 SOLV OW 80 -1.114253284 1.125814730 -0.130726335
3 SOLV HW1 81 -1.050867782 1.164987609 -0.064034927
3 SOLV HW2 82 -1.062837217 1.074528079 -0.199473394
4 SOLV OW 83 1.470653724 -0.136787060 -1.009224839
4 SOLV HW1 84 1.539469535 -0.088044157 -0.955466500
4 SOLV HW2 85 1.488937293 -0.235039184 -1.005524050
5 SOLV OW 86 1.216448808 -0.626960595 -1.012420962
5 SOLV HW1 87 1.257661249 -0.593192160 -0.927786518
5 SOLV HW2 88 1.233101950 -0.725225787 -1.020688434
6 SOLV OW 89 0.320401420 1.559303182 0.955261739
6 SOLV HW1 90 0.412194365 1.528579333 0.930128771
6 SOLV HW2 91 0.252544521 1.497491756 0.915558468
7 SOLV OW 92 0.949419785 -0.351062113 -0.141976508
7 SOLV HW1 93 0.912114289 -0.443850822 -0.142188122
7 SOLV HW2 94 1.021249331 -0.343874448 -0.211188637
8 SOLV OW 95 0.958787753 -0.914270034 -1.168159611
8 SOLV HW1 96 1.032683645 -0.979204522 -1.186171666
8 SOLV HW2 97 0.928700965 -0.923527880 -1.073235766
9 SOLV OW 98 -1.233115694 0.027205044 -1.263183241
9 SOLV HW1 99 -1.276112057 -0.044576653 -1.317960619
9 SOLV HW2 100 -1.190644308 -0.013529879 -1.182320781
10 SOLV OW 101 0.151358093 -1.343645803 -0.647417945
10 SOLV HW1 102 0.141455363 -1.291554920 -0.732212662
10 SOLV HW2 103 0.117371098 -1.289209596 -0.570715753
11 SOLV OW 104 -0.589462246 0.544235023 0.463474587
11 SOLV HW1 105 -0.502675232 0.511558037 0.426032914
11 SOLV HW2 106 -0.604889404 0.502564829 0.553069087
12 SOLV OW 107 0.996917417 0.281229442 0.391238737
12 SOLV HW1 108 0.960037272 0.370473978 0.365222919
12 SOLV HW2 109 1.017165981 0.228251213 0.308868684
13 SOLV OW 110 -0.518074322 -0.456930471 -1.364574820
13 SOLV HW1 111 -0.516607540 -0.413282379 -1.454542748
13 SOLV HW2 112 -0.549096757 -0.391009722 -1.296065042
14 SOLV OW 113 -0.896430622 1.529702824 0.657723813
14 SOLV HW1 114 -0.957865873 1.451186790 0.649801880
14 SOLV HW2 115 -0.801898986 1.497582756 0.663533171
15 SOLV OW 116 -0.757040297 -1.282591746 -0.598531405
15 SOLV HW1 117 -0.694156674 -1.204898512 -0.595218001
15 SOLV HW2 118 -0.851142986 -1.249642936 -0.590741587
16 SOLV OW 119 -0.505804702 -1.350350162 -1.311300259
16 SOLV HW1 120 -0.469537490 -1.424550826 -1.367683594
16 SOLV HW2 121 -0.507543248 -1.265780780 -1.364639618
17 SOLV OW 122 -1.102976732 -0.726997997 1.029523503
17 SOLV HW1 123 -1.114506045 -0.751301624 1.125846881
17 SOLV HW2 124 -1.023785459 -0.775354084 0.992212835
18 SOLV OW 125 1.270326808 1.127053927 -0.049953716
18 SOLV HW1 126 1.337740101 1.098081858 -0.117909869
18 SOLV HW2 127 1.272742300 1.064577376 0.028102251
19 SOLV OW 128 -1.355839933 -1.026886748 1.221881469
19 SOLV HW1 129 -1.378633044 -1.097070120 1.289385459
19 SOLV HW2 130 -1.437899025 -0.974268872 1.199528900
20 SOLV OW 131 -0.095658762 -1.054950191 0.431231590
20 SOLV HW1 132 -0.003486993 -1.085108252 0.455658904
20 SOLV HW2 133 -0.152800079 -1.052666103 0.513277592
21 SOLV OW 134 1.444330332 -1.075334691 -1.224115400
21 SOLV HW1 135 1.391170155 -1.147961105 -1.180515350
21 SOLV HW2 136 1.541552843 -1.089341023 -1.205325367
22 SOLV OW 137 -0.929831158 -0.190609585 0.300858977
22 SOLV HW1 138 -0.934773171 -0.242591065 0.215564991
22 SOLV HW2 139 -0.853929695 -0.125675706 0.295951151
23 SOLV OW 140 0.782604019 -0.182038547 -0.889598041
23 SOLV HW1 141 0.742462942 -0.258719924 -0.839494634
23 SOLV HW2 142 0.823572979 -0.215493493 -0.974474044
24 SOLV OW 143 -0.825777234 -0.258468951 0.588644965
24 SOLV HW1 144 -0.905928712 -0.221674958 0.541491418
24 SOLV HW2 145 -0.847678616 -0.348496452 0.626285514
25 SOLV OW 146 -1.067514593 -0.191834224 0.940104706
25 SOLV HW1 147 -1.047387444 -0.225597609 0.848145369
25 SOLV HW2 148 -1.143452207 -0.244083451 0.978899484
26 SOLV OW 149 -1.511131810 -0.786646194 1.442582219
26 SOLV HW1 150 -1.522452803 -0.853227003 1.368821172
26 SOLV HW2 151 -1.577966966 -0.805090973 1.514656367
27 SOLV OW 152 1.055269316 -0.444633549 0.598479279
27 SOLV HW1 153 1.011273715 -0.518212094 0.546976928
27 SOLV HW2 154 1.148417676 -0.471920190 0.622577178
28 SOLV OW 155 -0.326284749 0.443850524 0.487926666
28 SOLV HW1 156 -0.280691384 0.397848958 0.411734595
28 SOLV HW2 157 -0.341197868 0.378525449 0.562158220
29 SOLV OW 158 -1.178733145 0.333752360 -0.472395511
29 SOLV HW1 159 -1.134183431 0.403816566 -0.528147023
29 SOLV HW2 160 -1.236695401 0.276726135 -0.530621333
30 SOLV OW 161 0.597556320 -0.838582085 0.529788956
30 SOLV HW1 162 0.577023663 -0.771736968 0.458292625
30 SOLV HW2 163 0.583291803 -0.796819346 0.619532783
31 SOLV OW 164 0.092987249 0.900887980 -1.178459214
31 SOLV HW1 165 0.178494885 0.864505930 -1.141494538
31 SOLV HW2 166 0.104259207 0.919854286 -1.276002631
32 SOLV OW 167 0.671849257 -1.226849793 0.071569823
32 SOLV HW1 168 0.588152613 -1.205776915 0.122090454
32 SOLV HW2 169 0.687258461 -1.156763823 0.001913287
33 SOLV OW 170 0.693031585 -0.080532995 0.309431114
33 SOLV HW1 171 0.647872647 -0.167634270 0.328823710
33 SOLV HW2 172 0.769623625 -0.067954272 0.372497516
34 SOLV OW 173 -0.959083044 1.294222355 -1.047394358
34 SOLV HW1 174 -0.944998079 1.368170146 -0.981551550
34 SOLV HW2 175 -0.907581109 1.313698207 -1.130881996
35 SOLV OW 176 0.270807150 1.085028389 0.138368528
35 SOLV HW1 177 0.231069735 1.159639073 0.084942761
35 SOLV HW2 178 0.237116138 0.997535365 0.103583812
36 SOLV OW 179 -0.210996912 0.752801116 -0.604209241
36 SOLV HW1 180 -0.185858762 0.814969487 -0.530013597
36 SOLV HW2 181 -0.194868459 0.797630149 -0.692140019
37 SOLV OW 182 0.507632092 0.991858161 -1.060188897
37 SOLV HW1 183 0.560047876 0.966099276 -0.979005618
37 SOLV HW2 184 0.453820666 1.073818906 -1.040479716
38 SOLV OW 185 -1.136101540 1.257138515 -1.512036347
38 SOLV HW1 186 -1.135949427 1.292614477 -1.418540017
38 SOLV HW2 187 -1.229792093 1.235032563 -1.539119500
39 SOLV OW 188 -0.037975330 -0.525706592 0.927678025
39 SOLV HW1 189 0.056171240 -0.559271401 0.924292340
39 SOLV HW2 190 -0.060645100 -0.499491395 1.021488519
40 SOLV OW 191 -1.201463694 0.634869748 -1.025823916
40 SOLV HW1 192 -1.145438718 0.572498174 -0.971302489
40 SOLV HW2 193 -1.159654706 0.647157531 -1.115838609
41 SOLV OW 194 0.817219897 -1.194166184 -0.976779996
41 SOLV HW1 195 0.772697081 -1.231507948 -0.895384820
41 SOLV HW2 196 0.899060084 -1.143471457 -0.949687413
42 SOLV OW 197 1.161498550 -0.524017917 -0.391619303
42 SOLV HW1 198 1.147788632 -0.605224692 -0.448357472
42 SOLV HW2 199 1.101187845 -0.450752826 -0.423185801
43 SOLV OW 200 -0.530877617 1.430818531 1.121273148
43 SOLV HW1 201 -0.558054793 1.479119652 1.204510974
43 SOLV HW2 202 -0.478032080 1.349510274 1.145694586
44 SOLV OW 203 -0.881169676 -0.498127985 0.680169246
44 SOLV HW1 204 -0.841091625 -0.586243923 0.655052242
44 SOLV HW2 205 -0.958484683 -0.513054331 0.741821806
45 SOLV OW 206 -0.226684445 0.988191851 -1.041549808
45 SOLV HW1 207 -0.190589363 1.061165616 -0.983466545
45 SOLV HW2 208 -0.183833256 0.993155613 -1.131776350
46 SOLV OW 209 -0.156935800 0.534855376 0.825718599
46 SOLV HW1 210 -0.215623360 0.576029963 0.755987264
46 SOLV HW2 211 -0.063363480 0.568328865 0.814505860
47 SOLV OW 212 -0.707012048 1.143690438 0.608771382
47 SOLV HW1 213 -0.682986844 1.127486560 0.704481126
47 SOLV HW2 214 -0.786919201 1.203639203 0.604179796
48 SOLV OW 215 -1.529802166 1.212808446 -0.618249816
48 SOLV HW1 216 -1.581781496 1.167332199 -0.545916372
48 SOLV HW2 217 -1.450105359 1.258269248 -0.578453307
49 SOLV OW 218 1.418463146 1.225784050 1.477606300
49 SOLV HW1 219 1.518414483 1.228087707 1.475164237
49 SOLV HW2 220 1.388193511 1.143167951 1.525143605
50 SOLV OW 221 1.109154861 0.524552775 -0.461692954
50 SOLV HW1 222 1.169360910 0.594252864 -0.422722016
50 SOLV HW2 223 1.014095827 0.546647687 -0.439848160
51 SOLV OW 224 0.703752531 -0.505458479 0.624399001
51 SOLV HW1 225 0.660864726 -0.430714753 0.573662977
51 SOLV HW2 226 0.757938874 -0.467892799 0.699583774
52 SOLV OW 227 0.214631257 -0.805790517 0.425743786
52 SOLV HW1 228 0.277162399 -0.761837721 0.490241253
52 SOLV HW2 229 0.131432994 -0.750930556 0.417366168
53 SOLV OW 230 1.222026330 0.432029546 0.690036494
53 SOLV HW1 231 1.263063625 0.348836018 0.652666482
53 SOLV HW2 232 1.294333354 0.495779429 0.716671498
54 SOLV OW 233 0.682327505 1.509551780 -0.798271516
54 SOLV HW1 234 0.597377931 1.559068753 -0.780015467
54 SOLV HW2 235 0.731855906 1.495617682 -0.712514063
55 SOLV OW 236 0.505300403 1.179740329 1.516857890
55 SOLV HW1 237 0.434005394 1.229188305 1.566596421
55 SOLV HW2 238 0.478766447 1.083796186 1.507229900
56 SOLV OW 239 -0.962837345 -1.269003263 0.655655481
56 SOLV HW1 240 -0.960978155 -1.219830913 0.568591585
56 SOLV HW2 241 -0.946329094 -1.204803144 0.730537178
57 SOLV OW 242 0.862664526 -1.101334022 1.272159900
57 SOLV HW1 243 0.909593700 -1.157702272 1.204186647
57 SOLV HW2 244 0.930124210 -1.050341126 1.325518262
58 SOLV OW 245 -1.418026067 1.404692677 -0.983433323
58 SOLV HW1 246 -1.464970410 1.316667936 -0.976378075
58 SOLV HW2 247 -1.417089356 1.434386626 -1.078928154
59 SOLV OW 248 -1.145130813 0.036438001 -0.425237755
59 SOLV HW1 249 -1.238552389 0.068034769 -0.441843001
59 SOLV HW2 250 -1.080360758 0.103030071 -0.462276807
60 SOLV OW 251 -0.593636302 -0.687528281 -0.733431175
60 SOLV HW1 252 -0.646874583 -0.707916472 -0.815600163
60 SOLV HW2 253 -0.553950515 -0.596068803 -0.741289865
61 SOLV OW 254 1.286787252 1.287388716 0.676385801
61 SOLV HW1 255 1.279183265 1.289743137 0.776069143
61 SOLV HW2 256 1.253562560 1.373721085 0.638399856
62 SOLV OW 257 -0.191588629 -1.232054308 0.247552399
62 SOLV HW1 258 -0.151471640 -1.162153235 0.306764062
62 SOLV HW2 259 -0.205831141 -1.315992086 0.300021953
63 SOLV OW 260 0.214219754 -1.192088054 -1.363038072
63 SOLV HW1 261 0.216038465 -1.290864837 -1.378581405
63 SOLV HW2 262 0.179443948 -1.146137218 -1.444774769
64 SOLV OW 263 0.106241142 -0.178404690 1.378992492
64 SOLV HW1 264 0.057963891 -0.239121533 1.442115937
64 SOLV HW2 265 0.061394312 -0.089021805 1.377888935
65 SOLV OW 266 1.341373136 0.494893776 -0.009565136
65 SOLV HW1 267 1.414317560 0.432858038 0.019258572
65 SOLV HW2 268 1.253904172 0.446448921 -0.008026759
66 SOLV OW 269 1.050375926 0.379842145 1.550840648
66 SOLV HW1 270 1.091106954 0.288776538 1.543764350
66 SOLV HW2 271 1.018999963 0.409403698 1.460599008
67 SOLV OW 272 0.470519452 -0.726187470 0.297336841
67 SOLV HW1 273 0.374752434 -0.738273141 0.323499325
67 SOLV HW2 274 0.487440391 -0.629862406 0.276431918
68 SOLV OW 275 -0.546854363 1.128026755 -0.083826145
68 SOLV HW1 276 -0.487613443 1.088640468 -0.013532138
68 SOLV HW2 277 -0.593679479 1.208265687 -0.046797845
69 SOLV OW 278 0.742521201 -1.456708444 0.669055692
69 SOLV HW1 279 0.677273169 -1.382035480 0.656085460
69 SOLV HW2 280 0.832495697 -1.418912436 0.690911610
70 SOLV OW 281 -0.922949662 0.047006174 1.446402024
70 SOLV HW1 282 -0.830101851 0.029592395 1.413568732
70 SOLV HW2 283 -0.986160553 -0.017379001 1.403264680
71 SOLV OW 284 1.365356339 0.820404018 -1.102243309
71 SOLV HW1 285 1.396422144 0.904704428 -1.146178118
71 SOLV HW2 286 1.443495734 0.759733792 -1.087574398
72 SOLV OW 287 -0.966160695 0.365758172 1.283105615
72 SOLV HW1 288 -0.896398823 0.396007599 1.218142132
72 SOLV HW2 289 -0.923330740 0.346460695 1.371395452
73 SOLV OW 290 -1.285201437 0.134852992 0.189756503
73 SOLV HW1 291 -1.296321076 0.211727635 0.252738844
73 SOLV HW2 292 -1.221179498 0.160200240 0.117238930
74 SOLV OW 293 -0.831986044 0.493651687 -0.170858710
74 SOLV HW1 294 -0.810776222 0.466725978 -0.076915745
74 SOLV HW2 295 -0.863306440 0.413724953 -0.222150427
75 SOLV OW 296 -0.781661795 -0.129675118 -1.082082356
75 SOLV HW1 297 -0.722512476 -0.050402266 -1.096879139
75 SOLV HW2 298 -0.872470277 -0.098683228 -1.053885995
76 SOLV OW 299 0.353001346 1.566210164 -1.092605922
76 SOLV HW1 300 0.398373343 1.484680197 -1.128606763
76 SOLV HW2 301 0.342268721 1.557369192 -0.993568304
77 SOLV OW 302 0.868108415 1.249679117 0.736560730
77 SOLV HW1 303 0.881497783 1.294994286 0.648418533
77 SOLV HW2 304 0.940045643 1.181570662 0.750271533
78 SOLV OW 305 -1.513307840 0.953129002 1.254991780
78 SOLV HW1 306 -1.448363255 0.878907334 1.271573660
78 SOLV HW2 307 -1.595273606 0.916461545 1.210961500
79 SOLV OW 308 -1.213265052 0.797860395 0.515616198
79 SOLV HW1 309 -1.279479784 0.804997143 0.441009758
79 SOLV HW2 310 -1.137158749 0.860501715 0.498722576
80 SOLV OW 311 0.563442610 1.326911737 1.073031295
80 SOLV HW1 312 0.613526389 1.405257608 1.109823003
80 SOLV HW2 313 0.475016258 1.319276557 1.119123670
81 SOLV OW 314 -0.371247539 -1.082614839 1.304610380
81 SOLV HW1 315 -0.417326085 -1.090310715 1.393035732
81 SOLV HW2 316 -0.292726947 -1.021276356 1.313185147
82 SOLV OW 317 -1.385387780 -0.644210794 -1.408660102
82 SOLV HW1 318 -1.341600875 -0.563960913 -1.449191000
82 SOLV HW2 319 -1.315821410 -0.712687538 -1.386900465
83 SOLV OW 320 -0.242756717 1.040435022 -1.415915735
83 SOLV HW1 321 -0.259824843 0.941893841 -1.416464420
83 SOLV HW2 322 -0.190488357 1.065642867 -1.497368278
84 SOLV OW 323 -0.223222092 -0.324785141 -1.042589844
84 SOLV HW1 324 -0.161658113 -0.266952212 -0.989045337
84 SOLV HW2 325 -0.268469771 -0.390277490 -0.982048950
85 SOLV OW 326 1.501671426 0.844744366 -0.714933392
85 SOLV HW1 327 1.505482958 0.874457315 -0.619515459
85 SOLV HW2 328 1.566125558 0.769582831 -0.729017156
86 SOLV OW 329 1.395842524 0.565680194 -1.386308658
86 SOLV HW1 330 1.296675744 0.564688338 -1.373391164
86 SOLV HW2 331 1.417833052 0.532256875 -1.477966069
87 SOLV OW 332 -0.944301105 1.030771468 0.935741833
87 SOLV HW1 333 -0.940652522 0.975109319 0.852735086
87 SOLV HW2 334 -0.891001225 1.114303626 0.922208085
88 SOLV OW 335 -1.262091900 -0.231258203 0.156980864
88 SOLV HW1 336 -1.176011957 -0.255089271 0.111993633
88 SOLV HW2 337 -1.247823641 -0.226556023 0.255853772
89 SOLV OW 338 -0.817807703 -0.512316183 -0.118671460
89 SOLV HW1 339 -0.860602391 -0.462566658 -0.043203088
89 SOLV HW2 340 -0.767291288 -0.448189541 -0.176443999
90 SOLV OW 341 1.268163807 1.359186038 0.097332185
90 SOLV HW1 342 1.281043939 1.288884270 0.167283719
90 SOLV HW2 343 1.288189476 1.320623527 0.007257581
91 SOLV OW 344 -0.530238549 -0.265883213 0.232785864
91 SOLV HW1 345 -0.536187264 -0.262280255 0.133018885
91 SOLV HW2 346 -0.586529995 -0.193201998 0.272161286
92 SOLV OW 347 -0.350012166 0.542555907 -0.592516118
92 SOLV HW1 348 -0.421599530 0.551541010 -0.523261988
92 SOLV HW2 349 -0.298981940 0.628320088 -0.599003018
93 SOLV OW 350 1.279609320 -0.482022354 0.343897056
93 SOLV HW1 351 1.308557363 -0.396054479 0.386011299
93 SOLV HW2 352 1.280935572 -0.554811764 0.412468152
94 SOLV OW 353 0.946342794 0.754065436 0.059620429
94 SOLV HW1 354 1.006780782 0.777106375 0.135886115
94 SOLV HW2 355 0.948913909 0.827459906 -0.008267177
95 SOLV OW 356 -1.548741243 0.229800680 -0.416877205
95 SOLV HW1 357 -1.553199867 0.322746106 -0.380230723
95 SOLV HW2 358 -1.505480440 0.169572713 -0.349774662
96 SOLV OW 359 0.290873410 -0.505296574 0.043153056
96 SOLV HW1 360 0.353175912 -0.574304603 0.006299001
96 SOLV HW2 361 0.245727589 -0.457523949 -0.032222952
97 SOLV OW 362 0.075349363 -1.004608959 -1.143996709
97 SOLV HW1 363 0.133368880 -0.967925480 -1.216729711
97 SOLV HW2 364 0.043967587 -0.929807885 -1.085499905
98 SOLV OW 365 -0.250534541 1.190020256 0.277320324
98 SOLV HW1 366 -0.279976147 1.197751577 0.372584828
98 SOLV HW2 367 -0.156352758 1.156526394 0.274174737
99 SOLV OW 368 -0.768740916 0.066102282 0.656983366
99 SOLV HW1 369 -0.705664741 -0.011460471 0.654302433
99 SOLV HW2 370 -0.862873004 0.032418651 0.659497477
100 SOLV OW 371 1.253300657 1.399378894 -0.951886735
100 SOLV HW1 372 1.298897070 1.470497280 -0.898377209
100 SOLV HW2 373 1.294519796 1.394574424 -1.042870194
101 SOLV OW 374 1.103634300 1.224423757 1.429840365
101 SOLV HW1 375 1.057847732 1.137218625 1.412505642
101 SOLV HW2 376 1.179456373 1.210170986 1.493475360
102 SOLV OW 377 1.465854146 -0.060745536 -1.276593127
102 SOLV HW1 378 1.463210339 -0.095531464 -1.182864705
102 SOLV HW2 379 1.373419445 -0.060825296 -1.314773315
103 SOLV OW 380 -0.412582917 1.029177218 0.121290266
103 SOLV HW1 381 -0.468811397 1.052276404 0.200693514
103 SOLV HW2 382 -0.316973489 1.051249091 0.140623132
104 SOLV OW 383 -1.113479421 1.505089034 -0.061405516
104 SOLV HW1 384 -1.145060631 1.534997747 -0.151461912
104 SOLV HW2 385 -1.016254164 1.482135536 -0.066131468
105 SOLV OW 386 -1.273830498 0.874230816 -1.514400226
105 SOLV HW1 387 -1.296987253 0.818417620 -1.434712057
105 SOLV HW2 388 -1.199204218 0.936520831 -1.490898474
106 SOLV OW 389 -0.771485577 -0.089091271 -0.116167935
106 SOLV HW1 390 -0.737103858 -0.089881301 -0.022259508
106 SOLV HW2 391 -0.720553972 -0.155550725 -0.170854366
107 SOLV OW 392 1.096361500 -0.185928625 1.000349966
107 SOLV HW1 393 1.027019224 -0.239071957 1.049007620
107 SOLV HW2 394 1.054721833 -0.141604181 0.920967526
108 SOLV OW 395 1.147190380 0.882514173 -0.683461879
108 SOLV HW1 396 1.063039086 0.854038135 -0.729387745
108 SOLV HW2 397 1.224793923 0.832787587 -0.722275049
109 SOLV OW 398 1.010284067 -1.060002485 0.633076308
109 SOLV HW1 399 1.008612460 -1.076793886 0.534500578
109 SOLV HW2 400 1.032130004 -1.145027135 0.680985912
110 SOLV OW 401 0.015576758 0.832285473 -0.855056663
110 SOLV HW1 402 0.050479621 0.853063551 -0.946443265
110 SOLV HW2 403 -0.078757781 0.799818208 -0.862007307
111 SOLV OW 404 0.526090190 -0.207504699 1.159988621
111 SOLV HW1 405 0.478095234 -0.162022698 1.084959676
111 SOLV HW2 406 0.497082978 -0.167751877 1.247050532
112 SOLV OW 407 1.116903936 -1.164474863 0.983839594
112 SOLV HW1 408 1.186421363 -1.220524718 1.028870050
112 SOLV HW2 409 1.026031549 -1.194077586 1.013298889
113 SOLV OW 410 -0.571585161 1.114405830 0.851191884
113 SOLV HW1 411 -0.593582754 1.054472045 0.928170774
113 SOLV HW2 412 -0.473676464 1.134781351 0.851752838
114 SOLV OW 413 -1.308955592 0.319361180 0.393224991
114 SOLV HW1 414 -1.365579048 0.396788081 0.364930342
114 SOLV HW2 415 -1.226195668 0.353222446 0.438013259
115 SOLV OW 416 -0.803762783 -0.709726951 -0.537528260
115 SOLV HW1 417 -0.869135702 -0.777539461 -0.571113591
115 SOLV HW2 418 -0.737146114 -0.689304546 -0.609258552
116 SOLV OW 419 -1.248898574 -0.806057377 -0.264464858
116 SOLV HW1 420 -1.160422806 -0.759439591 -0.263810642
116 SOLV HW2 421 -1.234618808 -0.904482885 -0.274955144
117 SOLV OW 422 0.730338289 0.970150348 0.272746929
117 SOLV HW1 423 0.691737255 1.036573397 0.336773488
117 SOLV HW2 424 0.657849549 0.934953788 0.213518135
118 SOLV OW 425 0.580529555 0.883824294 -1.303544145
118 SOLV HW1 426 0.545848568 0.913875668 -1.214694642
118 SOLV HW2 427 0.650478106 0.947822740 -1.335378329
119 SOLV OW 428 0.548432566 0.668672046 -0.924555422
119 SOLV HW1 429 0.613892062 0.741059653 -0.946389781
119 SOLV HW2 430 0.538182295 0.608109184 -1.003477503
120 SOLV OW 431 0.373010984 -0.411054348 -1.508434468
120 SOLV HW1 432 0.280743378 -0.448786082 -1.500402102
120 SOLV HW2 433 0.404937997 -0.381642781 -1.418339458
121 SOLV OW 434 0.184485868 -1.191329338 0.495966898
121 SOLV HW1 435 0.265229660 -1.132419264 0.492484592
121 SOLV HW2 436 0.201817530 -1.274732103 0.443568729
122 SOLV OW 437 -0.274332385 -0.189048348 -1.281659973
122 SOLV HW1 438 -0.328247787 -0.231374683 -1.354485994
122 SOLV HW2 439 -0.262805677 -0.254143543 -1.206615398
123 SOLV OW 440 -0.025742177 -0.343824316 -1.460492215
123 SOLV HW1 441 0.030364289 -0.415799215 -1.501399472
123 SOLV HW2 442 -0.035137667 -0.360635331 -1.362355439
124 SOLV OW 443 1.563034418 -0.844016268 0.540437279
124 SOLV HW1 444 1.540694840 -0.746755565 0.546984449
124 SOLV HW2 445 1.491708083 -0.891164034 0.488561035
125 SOLV OW 446 -1.305061562 -1.095445456 0.158205729
125 SOLV HW1 447 -1.348436321 -1.162204016 0.218733013
125 SOLV HW2 448 -1.375182365 -1.036943180 0.117435552
126 SOLV OW 449 -0.639967209 -0.360562403 -1.112237005
126 SOLV HW1 450 -0.573617852 -0.350107318 -1.038142784
126 SOLV HW2 451 -0.687049507 -0.273628446 -1.127311420
127 SOLV OW 452 -1.221211093 -1.317026646 0.999726964
127 SOLV HW1 453 -1.199490030 -1.413531248 0.984997363
127 SOLV HW2 454 -1.218739563 -1.296896124 1.097658391
128 SOLV OW 455 -1.155818056 1.345318502 0.960252311
128 SOLV HW1 456 -1.126945070 1.254262970 0.930638699
128 SOLV HW2 457 -1.211192974 1.387669879 0.888547241
129 SOLV OW 458 0.540374594 0.506913459 -0.349305881
129 SOLV HW1 459 0.562893289 0.413845289 -0.320472643
129 SOLV HW2 460 0.590880209 0.529078355 -0.432720451
130 SOLV OW 461 -0.753133960 -0.709104185 0.591875912
130 SOLV HW1 462 -0.685338384 -0.764130953 0.543116800
130 SOLV HW2 463 -0.845032233 -0.739567299 0.566808682
131 SOLV OW 464 -0.369156007 1.319316898 -0.989407030
131 SOLV HW1 465 -0.275869197 1.299815009 -0.959091264
131 SOLV HW2 466 -0.433449684 1.262533002 -0.937990413
132 SOLV OW 467 -1.323557793 -1.331924937 -1.362781139
132 SOLV HW1 468 -1.254549767 -1.392711976 -1.323476166
132 SOLV HW2 469 -1.279924020 -1.269486388 -1.427583831
133 SOLV OW 470 -1.371092393 0.216985549 -1.070962301
133 SOLV HW1 471 -1.319501035 0.167071336 -1.140593091
133 SOLV HW2 472 -1.394864370 0.307686656 -1.105743366
134 SOLV OW 473 0.167117534 0.910785364 -0.035213598
134 SOLV HW1 474 0.105245734 0.834083924 -0.018174186
134 SOLV HW2 475 0.247618545 0.878262150 -0.084845550
135 SOLV OW 476 1.123207850 -0.382140300 1.421142829
135 SOLV HW1 477 1.076446550 -0.441097680 1.355271431
135 SOLV HW2 478 1.196158288 -0.331770467 1.374853665
136 SOLV OW 479 1.333091025 -0.695811688 0.522066148
136 SOLV HW1 480 1.294438760 -0.779608843 0.560591062
136 SOLV HW2 481 1.356516679 -0.632823877 0.596119268
137 SOLV OW 482 0.979028282 -0.970971635 -0.893373955
137 SOLV HW1 483 1.059550103 -1.007774210 -0.846877310
137 SOLV HW2 484 0.922289082 -0.921471486 -0.827567508
138 SOLV OW 485 -0.227307371 -0.232439726 0.057122049
138 SOLV HW1 486 -0.326358384 -0.246174000 0.058425451
138 SOLV HW2 487 -0.204045981 -0.152360991 0.112329063
139 SOLV OW 488 -0.924879641 0.580474820 -1.408769376
139 SOLV HW1 489 -0.947129605 0.625514366 -1.322293454
139 SOLV HW2 490 -0.837671602 0.532352156 -1.399795403
140 SOLV OW 491 -0.201684726 -0.830454565 1.306409967
140 SOLV HW1 492 -0.193346249 -0.783083436 1.394082859
140 SOLV HW2 493 -0.260643934 -0.777495060 1.245408667
141 SOLV OW 494 -1.499710665 -1.529798248 -0.837221305
141 SOLV HW1 495 -1.556207155 -1.558271201 -0.914673922
141 SOLV HW2 496 -1.432707036 -1.462202516 -0.867926388
142 SOLV OW 497 -0.861037522 -0.035693351 1.086584859
142 SOLV HW1 498 -0.777993791 -0.055761657 1.034599258
142 SOLV HW2 499 -0.940831887 -0.067984395 1.035677626
143 SOLV OW 500 -1.351372488 1.058044294 -0.777819127
143 SOLV HW1 501 -1.286151092 1.132762983 -0.790656829
143 SOLV HW2 502 -1.436663013 1.094349829 -0.740284267
144 SOLV OW 503 0.898563237 -0.024360889 0.473885389
144 SOLV HW1 504 0.986857550 -0.068310466 0.490444955
144 SOLV HW2 505 0.913224304 0.064583230 0.430576645
145 SOLV OW 506 0.333784906 -0.731686702 1.143488267
145 SOLV HW1 507 0.302466636 -0.683471953 1.061656572
145 SOLV HW2 508 0.255081908 -0.754807534 1.200698747
146 SOLV OW 509 0.022201283 -0.915295828 -0.303913420
146 SOLV HW1 510 0.030987147 -0.824149490 -0.344126854
146 SOLV HW2 511 -0.062749057 -0.957587209 -0.335483748
147 SOLV OW 512 1.169092996 1.122982240 -1.191701602
147 SOLV HW1 513 1.236458370 1.079471866 -1.131961487
147 SOLV HW2 514 1.080770476 1.126943548 -1.144973338
148 SOLV OW 515 0.470346793 0.837487392 0.241406664
148 SOLV HW1 516 0.448815880 0.886840295 0.157131590
148 SOLV HW2 517 0.485208046 0.902987609 0.315503552
149 SOLV OW 518 -1.220076822 -1.024138364 -1.290515179
149 SOLV HW1 519 -1.313860183 -1.027178717 -1.255914749
149 SOLV HW2 520 -1.212601367 -1.082143002 -1.371640215
150 SOLV OW 521 -0.960451577 -1.302230296 -0.015027194
150 SOLV HW1 522 -0.889999105 -1.327095175 -0.081507774
150 SOLV HW2 523 -1.050841495 -1.316174671 -0.055483328
151 SOLV OW 524 -1.500281067 -0.736976375 0.955386345
151 SOLV HW1 525 -1.580722127 -0.709972431 0.902469843
151 SOLV HW2 526 -1.445410664 -0.801362761 0.902059782
152 SOLV OW 527 0.016962594 1.314468568 1.117540187
152 SOLV HW1 528 -0.023210714 1.404987986 1.131413695
152 SOLV HW2 529 0.113718999 1.324189067 1.094220667
153 SOLV OW 530 0.503791482 0.146336825 -0.962732274
153 SOLV HW1 531 0.441780889 0.130479507 -1.039574774
153 SOLV HW2 532 0.507125202 0.064605565 -0.905196397
154 SOLV OW 533 1.192375329 -1.501739528 1.298141158
154 SOLV HW1 534 1.171814133 -1.593992482 1.330831367
154 SOLV HW2 535 1.120164502 -1.439110659 1.327554101
155 SOLV OW 536 -0.265403731 0.515635360 1.076918054
155 SOLV HW1 537 -0.312937497 0.603556837 1.080421737
155 SOLV HW2 538 -0.224048608 0.503699288 0.986645197
156 SOLV OW 539 -0.870334162 0.155054774 0.335344373
156 SOLV HW1 540 -0.947630780 0.187129279 0.390100179
156 SOLV HW2 541 -0.869656714 0.202813478 0.247478810
157 SOLV OW 542 -0.413905552 -0.114547383 0.817357236
157 SOLV HW1 543 -0.332355516 -0.062520946 0.842749292
157 SOLV HW2 544 -0.392854228 -0.212304939 0.818843956
158 SOLV OW 545 -0.000032647 -0.746606015 -1.011360251
158 SOLV HW1 546 0.048038929 -0.686599008 -0.947405482
158 SOLV HW2 547 -0.084623660 -0.779294985 -0.969197621
159 SOLV OW 548 -1.069813762 -0.237466369 -0.502796053
159 SOLV HW1 549 -1.084668121 -0.157772309 -0.444228697
159 SOLV HW2 550 -0.974539397 -0.266957883 -0.495385108
160 SOLV OW 551 -0.250091663 0.771234779 -0.899106266
160 SOLV HW1 552 -0.342704634 0.733552183 -0.901236963
160 SOLV HW2 553 -0.238835109 0.836236310 -0.974270488
161 SOLV OW 554 1.048311477 0.116215689 0.148394874
161 SOLV HW1 555 1.095378113 0.032935598 0.119226666
161 SOLV HW2 556 0.949481527 0.103170447 0.140393998
162 SOLV OW 557 1.270874308 0.768665738 0.509599445
162 SOLV HW1 558 1.222384288 0.855422936 0.520724061
162 SOLV HW2 559 1.206296851 0.697828458 0.481075874
163 SOLV OW 560 -0.363347233 0.336738569 1.245569285
163 SOLV HW1 561 -0.397405831 0.402211306 1.313060400
163 SOLV HW2 562 -0.318343496 0.386032351 1.171094937
164 SOLV OW 563 0.341857153 0.372223395 0.407574897
164 SOLV HW1 564 0.298172715 0.415634953 0.486369955
164 SOLV HW2 565 0.366724391 0.442155310 0.340547658
165 SOLV OW 566 0.552945383 -0.058409807 -0.781018395
165 SOLV HW1 567 0.554633870 -0.059888325 -0.681033653
165 SOLV HW2 568 0.643789427 -0.081016061 -0.816206540
166 SOLV OW 569 1.154392543 -1.302123283 -0.631404650
166 SOLV HW1 570 1.140910376 -1.213893077 -0.676519462
166 SOLV HW2 571 1.066390503 -1.336637768 -0.598754861
167 SOLV OW 572 0.418573258 -1.075989338 0.459957953
167 SOLV HW1 573 0.507341111 -1.029992175 0.462458558
167 SOLV HW2 574 0.377152821 -1.063585252 0.369779984
168 SOLV OW 575 0.561924685 -0.658770076 -0.560136295
168 SOLV HW1 576 0.513808585 -0.572965732 -0.578139212
168 SOLV HW2 577 0.495481608 -0.730329170 -0.538545345
169 SOLV OW 578 0.622421011 1.256081329 0.413448768
169 SOLV HW1 579 0.555912554 1.302214058 0.354724951
169 SOLV HW2 580 0.580983690 1.173871012 0.452519236
170 SOLV OW 581 -0.685382407 -1.389586399 -1.535069977
170 SOLV HW1 582 -0.769662777 -1.441769574 -1.521812104
170 SOLV HW2 583 -0.705350806 -1.291813828 -1.528449006
171 SOLV OW 584 0.981762244 -1.149115125 0.378381498
171 SOLV HW1 585 1.040644655 -1.104620691 0.310903810
171 SOLV HW2 586 0.955142723 -1.239508761 0.344907900
172 SOLV OW 587 -1.382788895 -0.830420661 -0.905952667
172 SOLV HW1 588 -1.386441648 -0.857662502 -0.809794453
172 SOLV HW2 589 -1.291193993 -0.847939398 -0.942082582
173 SOLV OW 590 -1.478400367 -0.435337489 0.379181941
173 SOLV HW1 591 -1.391718172 -0.403739105 0.417754842
173 SOLV HW2 592 -1.533058343 -0.478446124 0.450974987
174 SOLV OW 593 0.092103270 0.235978387 0.932972306
174 SOLV HW1 594 0.029591359 0.259286329 1.007477340
174 SOLV HW2 595 0.110613344 0.317616903 0.878249687
175 SOLV OW 596 -0.619136237 1.199973434 -1.513907477
175 SOLV HW1 597 -0.622304316 1.214920262 -1.415080923
175 SOLV HW2 598 -0.704272918 1.232292436 -1.555227179
176 SOLV OW 599 -0.724026830 1.239987685 -0.667367132
176 SOLV HW1 600 -0.669662064 1.229137843 -0.750606179
176 SOLV HW2 601 -0.771239825 1.328121925 -0.669668462
177 SOLV OW 602 -0.618309639 0.288878588 1.065456971
177 SOLV HW1 603 -0.547180588 0.277570360 1.134843115
177 SOLV HW2 604 -0.596992832 0.231651427 0.986259819
178 SOLV OW 605 -0.590936527 0.806387871 0.322337027
178 SOLV HW1 606 -0.581517751 0.716686561 0.365544848
178 SOLV HW2 607 -0.579292763 0.797114264 0.223441935
179 SOLV OW 608 1.340813279 -1.218127827 0.265158480
179 SOLV HW1 609 1.389170827 -1.194897903 0.349561267
179 SOLV HW2 610 1.283796740 -1.140902854 0.237099538
180 SOLV OW 611 0.548411799 -0.600958662 -1.219713713
180 SOLV HW1 612 0.534766308 -0.664633443 -1.295615155
180 SOLV HW2 613 0.516113299 -0.510100907 -1.246233332
181 SOLV OW 614 -0.514111317 1.152955266 -0.849749646
181 SOLV HW1 615 -0.484719051 1.170453533 -0.755773332
181 SOLV HW2 616 -0.559750425 1.064088618 -0.854381656
182 SOLV OW 617 -0.887081543 0.868456511 0.271682421
182 SOLV HW1 618 -0.791341966 0.841119846 0.262288910
182 SOLV HW2 619 -0.939566416 0.833140677 0.194225342
183 SOLV OW 620 0.714488334 0.906863679 -0.899658462
183 SOLV HW1 621 0.755842224 0.993028551 -0.929105418
183 SOLV HW2 622 0.778131954 0.858071037 -0.839906534
184 SOLV OW 623 0.689766597 -0.459951700 1.064874832
184 SOLV HW1 624 0.688028558 -0.550936013 1.023393159
184 SOLV HW2 625 0.643916318 -0.462958175 1.153703613
185 SOLV OW 626 -0.513435341 0.810414290 -1.487965936
185 SOLV HW1 627 -0.430417728 0.756498699 -1.502154447
185 SOLV HW2 628 -0.525413180 0.873847076 -1.564339382
186 SOLV OW 629 1.221058705 0.158822183 1.490100118
186 SOLV HW1 630 1.175847083 0.095617717 1.427150912
186 SOLV HW2 631 1.297035626 0.203886184 1.443215775
187 SOLV OW 632 1.350371726 -0.958774968 1.473850539
187 SOLV HW1 633 1.394524554 -1.008372252 1.399069082
187 SOLV HW2 634 1.363715365 -0.860485756 1.461094736
188 SOLV OW 635 -1.492688154 -0.525298686 1.423347440
188 SOLV HW1 636 -1.450270164 -0.492212857 1.507656570
188 SOLV HW2 637 -1.518624575 -0.621236848 1.434531416
189 SOLV OW 638 -1.235321237 1.187877663 0.746294858
189 SOLV HW1 639 -1.263576958 1.092433106 0.736600746
189 SOLV HW2 640 -1.301267770 1.247019866 0.699870100
190 SOLV OW 641 -0.783172871 -1.455696614 0.533603143
190 SOLV HW1 642 -0.808669036 -1.495647628 0.445536973
190 SOLV HW2 643 -0.852910596 -1.389753932 0.561709317
191 SOLV OW 644 -0.471722791 -0.357236150 1.124033830
191 SOLV HW1 645 -0.551055462 -0.356708983 1.184924771
191 SOLV HW2 646 -0.388001515 -0.350640833 1.178337532
192 SOLV OW 647 -1.274930088 -0.010629831 1.370633868
192 SOLV HW1 648 -1.289862162 -0.053612036 1.459692791
192 SOLV HW2 649 -1.221918794 -0.072253125 1.312373368
193 SOLV OW 650 1.298060028 -1.281622425 -1.104790554
193 SOLV HW1 651 1.350796354 -1.320715164 -1.180237728
193 SOLV HW2 652 1.210411879 -1.248030063 -1.139300226
194 SOLV OW 653 1.298764858 -0.686381642 1.331294516
194 SOLV HW1 654 1.273647076 -0.642186574 1.245170196
194 SOLV HW2 655 1.218230481 -0.730631077 1.370762293
195 SOLV OW 656 0.729391559 0.240372131 0.552774870
195 SOLV HW1 657 0.684372538 0.151317835 0.559418914
195 SOLV HW2 658 0.762096341 0.254278931 0.459294855
196 SOLV OW 659 0.989011620 -1.353954179 1.415850604
196 SOLV HW1 660 0.936612953 -1.290831782 1.358653582
196 SOLV HW2 661 0.926160407 -1.410629537 1.469134557
197 SOLV OW 662 -0.042805575 -0.209258665 -0.890469220
197 SOLV HW1 663 -0.079241435 -0.142485458 -0.825544078
197 SOLV HW2 664 0.049940868 -0.234971114 -0.863295102
198 SOLV OW 665 0.304420488 -0.273718692 -0.846742512
198 SOLV HW1 666 0.383631900 -0.243815252 -0.899972092
198 SOLV HW2 667 0.333228276 -0.295366648 -0.753449970
199 SOLV OW 668 -1.074405694 -1.252323740 1.499425724
199 SOLV HW1 669 -1.117522090 -1.168270966 1.532254752
199 SOLV HW2 670 -0.975382797 -1.246057685 1.511945853
200 SOLV OW 671 -1.398613160 1.498366208 0.256600102
200 SOLV HW1 672 -1.445628541 1.514259323 0.343416548
200 SOLV HW2 673 -1.460821488 1.518328727 0.180892033
201 SOLV OW 674 -1.506162090 0.130399520 1.374211183
201 SOLV HW1 675 -1.530491849 0.111697565 1.469394558
201 SOLV HW2 676 -1.420488639 0.083854792 1.351959990
202 SOLV OW 677 -0.689602450 -1.129671372 0.307727274
202 SOLV HW1 678 -0.736252572 -1.042264316 0.294166237
202 SOLV HW2 679 -0.622446379 -1.143235716 0.234883874
203 SOLV OW 680 -1.141932044 -0.396796127 1.218042912
203 SOLV HW1 681 -1.197343574 -0.474279344 1.187588982
203 SOLV HW2 682 -1.175792554 -0.364287489 1.306349873
204 SOLV OW 683 -0.317730110 -0.842178793 0.903899847
204 SOLV HW1 684 -0.355280280 -0.784374793 0.976357518
204 SOLV HW2 685 -0.267486767 -0.785415632 0.838669393
205 SOLV OW 686 1.089410802 -0.359498420 -0.743420106
205 SOLV HW1 687 1.035325042 -0.354371839 -0.827375929
205 SOLV HW2 688 1.094171766 -0.268933341 -0.701284732
206 SOLV OW 689 0.289222146 0.487895260 -1.422757515
206 SOLV HW1 690 0.297644741 0.450989357 -1.515325891
206 SOLV HW2 691 0.290718255 0.412820746 -1.356700594
207 SOLV OW 692 0.922672805 1.253432165 0.060780973
207 SOLV HW1 693 0.851305201 1.185106469 0.045290842
207 SOLV HW2 694 0.945609134 1.297871823 -0.025825591
208 SOLV OW 695 -0.837583347 0.098189243 -1.514622054
208 SOLV HW1 696 -0.923731849 0.141706484 -1.488425950
208 SOLV HW2 697 -0.846662100 -0.001092846 -1.506740119
209 SOLV OW 698 -0.998658212 0.978514454 -0.915244131
209 SOLV HW1 699 -1.039233492 0.982842543 -1.006550442
209 SOLV HW2 700 -0.974187047 0.883874173 -0.894118125
210 SOLV OW 701 0.547032539 -1.035699990 -1.546017090
210 SOLV HW1 702 0.552455293 -1.091577396 -1.463251974
210 SOLV HW2 703 0.639384280 -1.014205789 -1.577811085
211 SOLV OW 704 0.386323706 1.326303816 -0.871499179
211 SOLV HW1 705 0.340746405 1.318902678 -0.960201056
211 SOLV HW2 706 0.480573456 1.293864227 -0.879667052
212 SOLV OW 707 0.956180253 0.364141407 -0.936430903
212 SOLV HW1 708 0.996145668 0.421114543 -0.864606670
212 SOLV HW2 709 0.992779620 0.271324043 -0.929554955
213 SOLV OW 710 1.525529633 -0.369575348 -0.726580249
213 SOLV HW1 711 1.595507156 -0.425265884 -0.681818136
213 SOLV HW2 712 1.464279325 -0.428806387 -0.778944471
214 SOLV OW 713 -0.201720346 1.414670848 0.910259624
214 SOLV HW1 714 -0.290001295 1.442586442 0.948039579
214 SOLV HW2 715 -0.128076629 1.458448375 0.961836957
215 SOLV OW 716 0.724475123 -0.495831433 -0.411024893
215 SOLV HW1 717 0.672516778 -0.572320135 -0.449102976
215 SOLV HW2 718 0.737377214 -0.509917873 -0.312865846
216 SOLV OW 719 0.549848890 -0.167938015 -1.184603885
216 SOLV HW1 720 0.554159326 -0.186155338 -1.282845466
216 SOLV HW2 721 0.639475983 -0.137388885 -1.152424206
217 SOLV OW 722 -0.041838151 1.056549637 1.107328062
217 SOLV HW1 723 -0.035014748 1.155365622 1.121130713
217 SOLV HW2 724 0.030800517 1.010667322 1.158514625
218 SOLV OW 725 1.478278435 1.074296230 0.961350911
218 SOLV HW1 726 1.434747366 1.160300711 0.934735776
218 SOLV HW2 727 1.574250709 1.075442713 0.933242988
219 SOLV OW 728 0.243606687 -0.719414459 1.548719259
219 SOLV HW1 729 0.202921737 -0.748768191 1.462204826
219 SOLV HW2 730 0.264649641 -0.799664690 1.604563584
220 SOLV OW 731 1.305633733 0.541130668 1.477829442
220 SOLV HW1 732 1.343314571 0.539453896 1.570452085
220 SOLV HW2 733 1.320698132 0.452449170 1.434123518
221 SOLV OW 734 0.652564797 1.266215720 -0.382236122
221 SOLV HW1 735 0.667654804 1.179793284 -0.334226384
221 SOLV HW2 736 0.682383710 1.342127874 -0.324360078
222 SOLV OW 737 -1.431715214 1.335393026 0.627734216
222 SOLV HW1 738 -1.479360033 1.259129893 0.583984977
222 SOLV HW2 739 -1.446085552 1.418702315 0.574301383
223 SOLV OW 740 -0.726523267 -0.913467461 0.159673692
223 SOLV HW1 741 -0.630126037 -0.938825581 0.151511670
223 SOLV HW2 742 -0.734001639 -0.814508778 0.172054205
224 SOLV OW 743 -1.440659815 -0.105980201 -0.770494731
224 SOLV HW1 744 -1.518122938 -0.128970375 -0.711564828
224 SOLV HW2 745 -1.455281328 -0.015737177 -0.811043363
225 SOLV OW 746 1.107972446 1.420711125 -0.625485120
225 SOLV HW1 747 1.102403441 1.485483185 -0.549489312
225 SOLV HW2 748 1.197040656 1.375252301 -0.624053068
226 SOLV OW 749 -0.869381680 0.707919750 -0.853711873
226 SOLV HW1 750 -0.878409683 0.740679581 -0.759652923
226 SOLV HW2 751 -0.869582327 0.607913225 -0.854320639
227 SOLV OW 752 -0.480602634 -1.165063989 0.145459132
227 SOLV HW1 753 -0.471021600 -1.188272122 0.048653824
227 SOLV HW2 754 -0.400746143 -1.198201679 0.195723766
228 SOLV OW 755 1.398173109 0.546443614 0.962493384
228 SOLV HW1 756 1.400196377 0.578206379 0.867684604
228 SOLV HW2 757 1.406234893 0.446779204 0.964417421
229 SOLV OW 758 0.772431906 -0.139893042 -0.134398452
229 SOLV HW1 759 0.684352460 -0.159035488 -0.177724426
229 SOLV HW2 760 0.821237060 -0.225772613 -0.118770493
230 SOLV OW 761 -0.592512228 -0.861628470 -0.315495531
230 SOLV HW1 762 -0.667634182 -0.847589059 -0.250987108
230 SOLV HW2 763 -0.628727777 -0.897646139 -0.401477587
231 SOLV OW 764 0.192753748 -0.977499024 0.128163386
231 SOLV HW1 765 0.156263529 -0.938241700 0.212596815
231 SOLV HW2 766 0.131873508 -0.955192345 0.052020956
232 SOLV OW 767 -0.350398594 -0.338017773 -0.680220517
232 SOLV HW1 768 -0.309271184 -0.401184664 -0.745936346
232 SOLV HW2 769 -0.363708364 -0.384839296 -0.592855657
233 SOLV OW 770 -0.466128485 -1.276554042 -0.120810181
233 SOLV HW1 771 -0.373229940 -1.304682601 -0.144908175
233 SOLV HW2 772 -0.527779936 -1.354895797 -0.128791793
234 SOLV OW 773 -1.466771988 -0.197336017 0.004762741
234 SOLV HW1 774 -1.472117755 -0.262887784 -0.070577223
234 SOLV HW2 775 -1.388836908 -0.220014311 0.063187382
235 SOLV OW 776 -0.570620819 0.973994595 1.218431449
235 SOLV HW1 777 -0.575302054 1.033479853 1.298689427
235 SOLV HW2 778 -0.660537877 0.933562783 1.201641822
236 SOLV OW 779 0.088160947 1.057790960 -0.423581542
236 SOLV HW1 780 0.121143201 1.014275648 -0.507369531
236 SOLV HW2 781 0.021125997 0.998244955 -0.379282140
237 SOLV OW 782 0.991110815 -1.115835864 -0.269782387
237 SOLV HW1 783 1.050966331 -1.176168120 -0.217081094
237 SOLV HW2 784 0.895940965 -1.143690432 -0.256786730
238 SOLV OW 785 1.207094839 -0.679202659 -0.162886242
238 SOLV HW1 786 1.138550275 -0.750998478 -0.175072437
238 SOLV HW2 787 1.214311649 -0.625425252 -0.246894320
239 SOLV OW 788 0.031510836 -1.358066573 -0.086440327
239 SOLV HW1 789 -0.061212652 -1.332756211 -0.058806934
239 SOLV HW2 790 0.097136351 -1.321125273 -0.020634717
240 SOLV OW 791 -1.133803272 0.161100667 1.177183329
240 SOLV HW1 792 -1.057148606 0.223107926 1.193895340
240 SOLV HW2 793 -1.160180717 0.116889556 1.262913710
241 SOLV OW 794 0.907410053 0.759122047 -0.747565870
241 SOLV HW1 795 0.887824727 0.669604921 -0.707504554
241 SOLV HW2 796 0.963461041 0.747384062 -0.829555187
242 SOLV OW 797 -0.220091538 1.232037908 -0.390220188
242 SOLV HW1 798 -0.167603483 1.271072513 -0.465872329
242 SOLV HW2 799 -0.175799523 1.255078875 -0.303564497
243 SOLV OW 800 1.373523595 0.411921733 -0.437769967
243 SOLV HW1 801 1.284339303 0.371975052 -0.459033004
243 SOLV HW2 802 1.363473068 0.510476303 -0.424075105
244 SOLV OW 803 0.348063043 0.020269756 -1.154978929
244 SOLV HW1 804 0.275584959 0.009537201 -1.223050277
244 SOLV HW2 805 0.407765626 -0.059939866 -1.156959336
245 SOLV OW 806 -1.344665712 1.312101114 -0.190498580
245 SOLV HW1 807 -1.285043849 1.257179196 -0.131929996
245 SOLV HW2 808 -1.424546778 1.257989807 -0.216812736
246 SOLV OW 809 -1.024988868 0.950881477 -1.426453861
246 SOLV HW1 810 -0.936535959 0.927505326 -1.386061647
246 SOLV HW2 811 -1.010625472 0.992954234 -1.516038523
247 SOLV OW 812 -1.107417861 0.721019404 1.014631365
247 SOLV HW1 813 -1.008408924 0.728838790 1.002880340
247 SOLV HW2 814 -1.148892827 0.691012323 0.928716768
248 SOLV OW 815 -0.453341724 -0.926301014 0.270731026
248 SOLV HW1 816 -0.466131136 -1.007322201 0.213515429
248 SOLV HW2 817 -0.356096002 -0.903291107 0.274643200
249 SOLV OW 818 -0.461227710 0.821963137 -0.491136826
249 SOLV HW1 819 -0.381563550 0.817626376 -0.551438356
249 SOLV HW2 820 -0.509161981 0.908561632 -0.505435926
250 SOLV OW 821 0.541424875 -0.670584552 0.767655453
250 SOLV HW1 822 0.585712470 -0.686304662 0.855934465
250 SOLV HW2 823 0.587890126 -0.596016950 0.719882304
251 SOLV OW 824 -1.035301792 0.368632568 0.448316410
251 SOLV HW1 825 -0.980182816 0.346283789 0.528715569
251 SOLV HW2 826 -1.038539968 0.467909483 0.436691457
252 SOLV OW 827 1.460738309 -1.400370082 -1.298327078
252 SOLV HW1 828 1.439159962 -1.461857933 -1.374180553
252 SOLV HW2 829 1.546689203 -1.352921133 -1.317330531
253 SOLV OW 830 -0.752989986 -0.267585531 0.881637916
253 SOLV HW1 831 -0.704248966 -0.184355690 0.908073781
253 SOLV HW2 832 -0.762900388 -0.270213722 0.782156297
254 SOLV OW 833 -0.381036783 -0.525990590 0.434363971
254 SOLV HW1 834 -0.325754523 -0.551175146 0.354921270
254 SOLV HW2 835 -0.465171271 -0.481497012 0.403652487
255 SOLV OW 836 -0.604982595 1.067660831 0.345434570
255 SOLV HW1 837 -0.647634561 1.099885599 0.429957738
255 SOLV HW2 838 -0.601785167 0.967703262 0.345740953
256 SOLV OW 839 0.382151788 -1.462113066 1.506063279
256 SOLV HW1 840 0.404451163 -1.535541055 1.441933624
256 SOLV HW2 841 0.286953422 -1.471203697 1.535324144
257 SOLV OW 842 0.984559187 -0.980031319 1.479221572
257 SOLV HW1 843 1.033893596 -1.030704531 1.549932928
257 SOLV HW2 844 0.990246478 -0.882036517 1.498362057
258 SOLV OW 845 1.250299772 0.263605340 -1.467986152
258 SOLV HW1 846 1.300415269 0.332511615 -1.415636389
258 SOLV HW2 847 1.303647997 0.237620876 -1.548477547
259 SOLV OW 848 -0.032277697 -0.870082710 0.036009089
259 SOLV HW1 849 -0.035188073 -0.804642277 -0.039559579
259 SOLV HW2 850 -0.091296256 -0.948008105 0.014891452
260 SOLV OW 851 -1.194085948 1.242785361 -0.875655875
260 SOLV HW1 852 -1.123439959 1.257579009 -0.944880963
260 SOLV HW2 853 -1.276748804 1.292964161 -0.901164916
261 SOLV OW 854 0.694490128 0.668848861 -1.381177689
261 SOLV HW1 855 0.645798303 0.754638913 -1.364715323
261 SOLV HW2 856 0.721920766 0.628804053 -1.293738127
262 SOLV OW 857 0.690236772 -0.280450699 -1.400177368
262 SOLV HW1 858 0.762330266 -0.349704437 -1.397294664
262 SOLV HW2 859 0.691497333 -0.234733517 -1.489116159
263 SOLV OW 860 0.715940866 -0.737716865 1.441473842
263 SOLV HW1 861 0.760103843 -0.650219205 1.421585685
263 SOLV HW2 862 0.627833390 -0.721242702 1.485827367
264 SOLV OW 863 1.301829390 0.292619259 -1.053737008
264 SOLV HW1 864 1.393210914 0.275689654 -1.090654424
264 SOLV HW2 865 1.260172800 0.369696460 -1.101963918
265 SOLV OW 866 0.576369723 -0.998015068 -0.742437425
265 SOLV HW1 867 0.623100433 -1.082660991 -0.767997825
265 SOLV HW2 868 0.533000717 -1.009588213 -0.653066479
266 SOLV OW 869 1.307816868 -0.193832485 -0.160575677
266 SOLV HW1 870 1.265437150 -0.197735737 -0.251077172
266 SOLV HW2 871 1.291157855 -0.279793167 -0.112254164
267 SOLV OW 872 1.423643342 -1.112876046 -1.504309692
267 SOLV HW1 873 1.429988612 -1.066056813 -1.416174391
267 SOLV HW2 874 1.482133631 -1.193979709 -1.503227109
268 SOLV OW 875 -0.226595574 -1.059172055 0.703114796
268 SOLV HW1 876 -0.266189669 -0.993217396 0.767019682
268 SOLV HW2 877 -0.127052070 -1.059254369 0.712736624
269 SOLV OW 878 0.476670908 0.077211476 -1.512459159
269 SOLV HW1 879 0.448940084 0.124157649 -1.428620750
269 SOLV HW2 880 0.538467958 0.136001027 -1.564676981
270 SOLV OW 881 1.475830976 -0.404717487 -1.246916819
270 SOLV HW1 882 1.545317894 -0.440071930 -1.184279199
270 SOLV HW2 883 1.392414644 -0.383910257 -1.195823310
271 SOLV OW 884 0.418198932 -1.348402951 -0.321091479
271 SOLV HW1 885 0.379420852 -1.355524558 -0.229183105
271 SOLV HW2 886 0.441176178 -1.439739038 -0.354725946
272 SOLV OW 887 -0.221609562 -1.003194439 -0.396921538
272 SOLV HW1 888 -0.219540203 -1.099121545 -0.425122863
272 SOLV HW2 889 -0.276498304 -0.994077031 -0.313820443
273 SOLV OW 890 0.551537918 -1.059398929 -0.104139899
273 SOLV HW1 891 0.542482873 -0.971294596 -0.057693999
273 SOLV HW2 892 0.510375365 -1.053499120 -0.195092427
274 SOLV OW 893 1.491995921 -1.030406508 -0.516538535
274 SOLV HW1 894 1.414099157 -1.091215948 -0.501178802
274 SOLV HW2 895 1.464530757 -0.935918970 -0.498667614
275 SOLV OW 896 0.429959672 0.165566599 1.389915403
275 SOLV HW1 897 0.526342467 0.140839092 1.399939183
275 SOLV HW2 898 0.421232337 0.239002123 1.322591374
276 SOLV OW 899 -1.040678307 1.325672539 -0.523502585
276 SOLV HW1 900 -1.005762721 1.249511057 -0.578109322
276 SOLV HW2 901 -0.967986840 1.392997070 -0.509907043
277 SOLV OW 902 0.908836890 -0.357736791 -1.058511388
277 SOLV HW1 903 0.956135759 -0.287026535 -1.111094076
277 SOLV HW2 904 0.945967702 -0.447467422 -1.082421151
278 SOLV OW 905 0.748216854 -0.142756967 -0.503698287
278 SOLV HW1 906 0.780030518 -0.048538132 -0.514225922
278 SOLV HW2 907 0.814505810 -0.205119825 -0.545131828
279 SOLV OW 908 0.541969428 -0.461571433 0.249371283
279 SOLV HW1 909 0.548841415 -0.426668770 0.155903330
279 SOLV HW2 910 0.550733585 -0.385738729 0.313979119
280 SOLV OW 911 0.476709538 0.975062968 1.171582789
280 SOLV HW1 912 0.523764925 1.040267991 1.231043759
280 SOLV HW2 913 0.540227760 0.902649208 1.144690707
281 SOLV OW 914 0.805405809 0.028379530 -1.193255326
281 SOLV HW1 915 0.773475271 0.120125073 -1.169483606
281 SOLV HW2 916 0.790951030 0.012337706 -1.290905537
282 SOLV OW 917 -0.649714440 0.442973190 1.556127960
282 SOLV HW1 918 -0.735299358 0.394948263 1.575383337
282 SOLV HW2 919 -0.652382872 0.479838749 1.463199267
283 SOLV OW 920 1.080055644 0.759058735 0.920789569
283 SOLV HW1 921 1.077939759 0.659620970 0.931248159
283 SOLV HW2 922 1.165175589 0.794797814 0.959246994
284 SOLV OW 923 1.323958332 -1.303731890 1.164185607
284 SOLV HW1 924 1.362991863 -1.231790102 1.221650792
284 SOLV HW2 925 1.267969174 -1.364590663 1.220424778
285 SOLV OW 926 -1.494407069 -0.943390159 0.007201472
285 SOLV HW1 927 -1.577795253 -0.904944648 0.046826132
285 SOLV HW2 928 -1.518074977 -1.020659046 -0.051715588
286 SOLV OW 929 -0.381816895 1.522169418 0.543941116
286 SOLV HW1 930 -0.354504021 1.500109460 0.450295894
286 SOLV HW2 931 -0.422053770 1.441280677 0.586835301
287 SOLV OW 932 -0.126560994 -0.675618807 1.519193743
287 SOLV HW1 933 -0.090069817 -0.613102373 1.450185875
287 SOLV HW2 934 -0.208348603 -0.635554506 1.560516444
288 SOLV OW 935 1.011810906 -0.866489056 -0.160916014
288 SOLV HW1 936 0.998300070 -0.953927074 -0.207543263
288 SOLV HW2 937 0.983945766 -0.875096601 -0.065253015
289 SOLV OW 938 -0.104142896 -0.645847289 0.677033110
289 SOLV HW1 939 -0.092434155 -0.612222916 0.770488553
289 SOLV HW2 940 -0.016162340 -0.643566447 0.629538217
290 SOLV OW 941 1.291377146 0.034839453 1.032281737
290 SOLV HW1 942 1.256125071 0.127960072 1.041643268
290 SOLV HW2 943 1.215239854 -0.028257465 1.017326936
291 SOLV OW 944 -0.159777833 0.480578863 -0.385770094
291 SOLV HW1 945 -0.215815395 0.511705707 -0.309007995
291 SOLV HW2 946 -0.214704073 0.480752078 -0.469343398
292 SOLV OW 947 -0.711807780 -0.643769668 0.196858695
292 SOLV HW1 948 -0.694017050 -0.580629998 0.272347229
292 SOLV HW2 949 -0.808724684 -0.640065027 0.172465098
293 SOLV OW 950 0.594082904 -0.283879887 0.447597094
293 SOLV HW1 951 0.659748169 -0.236097344 0.505963525
293 SOLV HW2 952 0.504333676 -0.284162287 0.491719206
294 SOLV OW 953 0.724387852 -0.386381680 -0.719702445
294 SOLV HW1 954 0.728032496 -0.474063583 -0.767647107
294 SOLV HW2 955 0.673646103 -0.397248406 -0.634208478
295 SOLV OW 956 0.205808377 1.299210794 -0.032065932
295 SOLV HW1 957 0.259039791 1.383030647 -0.020201022
295 SOLV HW2 958 0.193776669 1.280927583 -0.129641811
296 SOLV OW 959 0.493343946 0.591472592 1.066073162
296 SOLV HW1 960 0.396868819 0.584985540 1.040535495
296 SOLV HW2 961 0.524754811 0.685643254 1.053951947
297 SOLV OW 962 -1.226121714 -0.325968450 -0.697900030
297 SOLV HW1 963 -1.307364653 -0.267648303 -0.697420327
297 SOLV HW2 964 -1.155365926 -0.285001800 -0.640305993
298 SOLV OW 965 -1.492294569 -0.712413793 -0.154498273
298 SOLV HW1 966 -1.474049638 -0.772335279 -0.076535965
298 SOLV HW2 967 -1.411428371 -0.708583510 -0.213214759
299 SOLV OW 968 -0.621473218 0.117160483 -1.064989450
299 SOLV HW1 969 -0.530277691 0.083985753 -1.040816664
299 SOLV HW2 970 -0.622929647 0.143253614 -1.161521840
300 SOLV OW 971 -0.062909076 1.313206268 -0.600107241
300 SOLV HW1 972 -0.072332013 1.406946968 -0.633656240
300 SOLV HW2 973 0.025877025 1.276601487 -0.628013160
301 SOLV OW 974 1.109396460 0.768093081 1.468485449
301 SOLV HW1 975 1.189410891 0.708412713 1.462361495
301 SOLV HW2 976 1.068882707 0.760256858 1.559582612
302 SOLV OW 977 -0.859613845 0.277461903 -1.159246799
302 SOLV HW1 978 -0.816605488 0.356132153 -1.203534577
302 SOLV HW2 979 -0.790792174 0.226659374 -1.107451329
303 SOLV OW 980 -0.175243825 -0.601860141 -0.748601552
303 SOLV HW1 981 -0.187638917 -0.692812872 -0.788296658
303 SOLV HW2 982 -0.077755198 -0.584776243 -0.734255387
304 SOLV OW 983 -1.507795316 -0.786422272 0.432874376
304 SOLV HW1 984 -1.411693071 -0.791381953 0.405641339
304 SOLV HW2 985 -1.549056584 -0.877274295 0.426149259
305 SOLV OW 986 -0.329073023 1.113608726 1.489777767
305 SOLV HW1 987 -0.421210649 1.143692419 1.514427679
305 SOLV HW2 988 -0.306682146 1.030166860 1.540156524
306 SOLV OW 989 -1.207777603 0.622366710 0.776183257
306 SOLV HW1 990 -1.225497540 0.652857189 0.682598684
306 SOLV HW2 991 -1.183381298 0.525381693 0.775566550
307 SOLV OW 992 1.566659812 1.071046522 0.260118863
307 SOLV HW1 993 1.568744780 1.144847905 0.192659205
307 SOLV HW2 994 1.477786682 1.070330199 0.305975311
308 SOLV OW 995 -0.456289850 -1.272522937 1.095158252
308 SOLV HW1 996 -0.386088128 -1.331886125 1.055793287
308 SOLV HW2 997 -0.541643136 -1.323742170 1.104814911
309 SOLV OW 998 -1.101498589 -0.466740895 -0.890817494
309 SOLV HW1 999 -1.131339284 -0.428078804 -0.803554076
309 SOLV HW2 1000 -1.002617216 -0.455209047 -0.900282204
310 SOLV OW 1001 0.663749416 -1.219727718 0.913681730
310 SOLV HW1 1002 0.578777424 -1.225453057 0.861254598
310 SOLV HW2 1003 0.657917404 -1.143690889 0.978381091
311 SOLV OW 1004 -1.561532638 0.420614684 0.967680781
311 SOLV HW1 1005 -1.558524452 0.502190770 0.909906174
311 SOLV HW2 1006 -1.469290919 0.399100584 0.999776046
312 SOLV OW 1007 -1.126326662 -0.009779250 0.378043105
312 SOLV HW1 1008 -1.178975926 0.036636379 0.306801353
312 SOLV HW2 1009 -1.033840874 -0.027961198 0.344613702
313 SOLV OW 1010 0.113800696 0.756740844 -0.428947925
313 SOLV HW1 1011 0.122642831 0.810381314 -0.512889857
313 SOLV HW2 1012 0.045432940 0.684984880 -0.442312444
314 SOLV OW 1013 -1.329275267 -0.575310519 -1.007016772
314 SOLV HW1 1014 -1.233289615 -0.559740807 -0.983683774
314 SOLV HW2 1015 -1.359995228 -0.661228878 -0.966072921
315 SOLV OW 1016 -0.664586827 0.903247710 -0.859325222
315 SOLV HW1 1017 -0.596358213 0.850064295 -0.909508437
315 SOLV HW2 1018 -0.754417532 0.860565700 -0.869837589
316 SOLV OW 1019 0.871692415 -0.774780942 1.194278189
316 SOLV HW1 1020 0.857988886 -0.865169058 1.153752479
316 SOLV HW2 1021 0.826961467 -0.771219430 1.283645678
317 SOLV OW 1022 -0.362936114 -0.726352504 -0.385750333
317 SOLV HW1 1023 -0.446531484 -0.781118208 -0.382192015
317 SOLV HW2 1024 -0.295753668 -0.771880230 -0.444178368
318 SOLV OW 1025 -0.558775842 -0.622181452 -0.019659108
318 SOLV HW1 1026 -0.612282263 -0.565593338 -0.082388509
318 SOLV HW2 1027 -0.599283763 -0.618092324 0.071678056
319 SOLV OW 1028 -1.515539268 1.162410046 -1.004566491
319 SOLV HW1 1029 -1.497955165 1.138301878 -1.100019793
319 SOLV HW2 1030 -1.523446464 1.078947627 -0.950039560
320 SOLV OW 1031 -0.475446390 0.376942209 -1.218518935
320 SOLV HW1 1032 -0.441332654 0.426920187 -1.138892838
320 SOLV HW2 1033 -0.400189096 0.326623834 -1.261018401
321 SOLV OW 1034 0.503840862 -0.289620674 1.567558085
321 SOLV HW1 1035 0.509637839 -0.384843632 1.537540987
321 SOLV HW2 1036 0.448669960 -0.237471287 1.502453757
322 SOLV OW 1037 0.735721459 -0.572604646 -0.913170459
322 SOLV HW1 1038 0.695416226 -0.654541281 -0.953961365
322 SOLV HW2 1039 0.794543526 -0.527468329 -0.980287156
323 SOLV OW 1040 -1.160173951 -0.788632146 -1.429322876
323 SOLV HW1 1041 -1.086847061 -0.721919466 -1.416180662
323 SOLV HW2 1042 -1.135945885 -0.874197680 -1.383588530
324 SOLV OW 1043 -0.755864496 -0.450530586 0.396495400
324 SOLV HW1 1044 -0.768566545 -0.361829687 0.440907879
324 SOLV HW2 1045 -0.799985835 -0.521753558 0.451105303
325 SOLV OW 1046 1.076709271 -1.184330096 -1.271893259
325 SOLV HW1 1047 1.072502373 -1.281632662 -1.294614839
325 SOLV HW2 1048 0.992405069 -1.139499207 -1.301636711
326 SOLV OW 1049 0.011174007 0.955521638 0.264060557
326 SOLV HW1 1050 -0.004924516 0.880315596 0.200146386
326 SOLV HW2 1051 0.058165427 1.030006783 0.216691161
327 SOLV OW 1052 -1.133343536 0.928095785 -1.147582411
327 SOLV HW1 1053 -1.088488396 0.976383265 -1.222791901
327 SOLV HW2 1054 -1.210403383 0.982474024 -1.114342717
328 SOLV OW 1055 -0.059206827 -0.126077368 1.121483069
328 SOLV HW1 1056 0.029947795 -0.158241489 1.089563545
328 SOLV HW2 1057 -0.053058337 -0.029156188 1.145364721
329 SOLV OW 1058 0.438230021 -1.074362868 0.853155162
329 SOLV HW1 1059 0.447213516 -0.987674572 0.804119192
329 SOLV HW2 1060 0.367852934 -1.065607103 0.923671061
330 SOLV OW 1061 1.448773701 -0.524729004 -0.380412162
330 SOLV HW1 1062 1.351386909 -0.503646445 -0.371871848
330 SOLV HW2 1063 1.498939738 -0.481357910 -0.305553016
331 SOLV OW 1064 -1.143182310 1.423811434 1.356832411
331 SOLV HW1 1065 -1.237484530 1.409406206 1.326808331
331 SOLV HW2 1066 -1.140829634 1.431999782 1.456477777
332 SOLV OW 1067 -0.833387623 1.394758304 0.357803984
332 SOLV HW1 1068 -0.776572984 1.328900515 0.308441369
332 SOLV HW2 1069 -0.791215393 1.485314634 0.353017450
333 SOLV OW 1070 0.313130720 0.840160301 -0.253351449
333 SOLV HW1 1071 0.237663729 0.803445016 -0.307746242
333 SOLV HW2 1072 0.361999534 0.909303629 -0.306577661
334 SOLV OW 1073 -0.674266368 0.470144046 -1.415035113
334 SOLV HW1 1074 -0.640664509 0.445343048 -1.505906443
334 SOLV HW2 1075 -0.598854807 0.466368951 -1.349456246
335 SOLV OW 1076 0.324011434 0.251722919 -1.309397758
335 SOLV HW1 1077 0.368253376 0.222828780 -1.224489741
335 SOLV HW2 1078 0.252063334 0.186517399 -1.333339002
336 SOLV OW 1079 1.383445479 1.297101910 -0.695255787
336 SOLV HW1 1080 1.407424878 1.217884920 -0.751392157
336 SOLV HW2 1081 1.423227152 1.379626141 -0.735365013
337 SOLV OW 1082 -0.653665070 1.058988781 1.455001868
337 SOLV HW1 1083 -0.656012672 1.092287571 1.549276165
337 SOLV HW2 1084 -0.745616203 1.063176497 1.415894920
338 SOLV OW 1085 -0.537405521 -0.590287723 0.953667210
338 SOLV HW1 1086 -0.590731346 -0.505931025 0.960149089
338 SOLV HW2 1087 -0.496959367 -0.597207640 0.862464908
339 SOLV OW 1088 0.921017012 0.878140342 1.117186389
339 SOLV HW1 1089 0.993462700 0.835251378 1.063203978
339 SOLV HW2 1090 0.922033405 0.976948271 1.101761082
340 SOLV OW 1091 -0.400582439 -1.028869108 -0.174358484
340 SOLV HW1 1092 -0.465782950 -0.980659088 -0.232880439
340 SOLV HW2 1093 -0.431325887 -1.123053064 -0.160782289
341 SOLV OW 1094 0.654429907 -0.843854634 -0.328152802
341 SOLV HW1 1095 0.737020784 -0.894872609 -0.304113599
341 SOLV HW2 1096 0.664043492 -0.806462413 -0.420409583
342 SOLV OW 1097 -0.009251142 1.514738476 -0.213242369
342 SOLV HW1 1098 0.052899077 1.437029340 -0.223271153
342 SOLV HW2 1099 -0.006161388 1.570764692 -0.296027739
343 SOLV OW 1100 0.852884689 -1.433607396 -0.741367239
343 SOLV HW1 1101 0.913430222 -1.415398327 -0.663879349
343 SOLV HW2 1102 0.859689278 -1.530029759 -0.767021725
344 SOLV OW 1103 0.027450928 -0.252630138 0.567319618
344 SOLV HW1 1104 0.073838631 -0.304501173 0.495490467
344 SOLV HW2 1105 -0.070207617 -0.274153735 0.566176501
345 SOLV OW 1106 0.082666662 0.618246313 -1.336513154
345 SOLV HW1 1107 0.162709469 0.569277758 -1.371113653
345 SOLV HW2 1108 0.111247144 0.683202469 -1.266044185
346 SOLV OW 1109 -0.848706518 0.503491357 -0.658580709
346 SOLV HW1 1110 -0.804618887 0.476625643 -0.572927456
346 SOLV HW2 1111 -0.789765028 0.478875921 -0.735534661
347 SOLV OW 1112 -0.749736548 1.413573088 1.359248979
347 SOLV HW1 1113 -0.786682515 1.334125908 1.311029080
347 SOLV HW2 1114 -0.775555003 1.497045490 1.310590449
348 SOLV OW 1115 0.966236820 1.175985047 -0.491588799
348 SOLV HW1 1116 1.017485894 1.210128094 -0.570389210
348 SOLV HW2 1117 1.014392713 1.200406882 -0.407408432
349 SOLV OW 1118 -1.463432401 1.021144179 1.541526877
349 SOLV HW1 1119 -1.393972692 1.092762889 1.548452836
349 SOLV HW2 1120 -1.496845229 1.016135370 1.447397068
350 SOLV OW 1121 -0.971396470 0.300696277 -0.278374319
350 SOLV HW1 1122 -0.919115161 0.220517488 -0.249399105
350 SOLV HW2 1123 -1.056834956 0.271099642 -0.321103411
351 SOLV OW 1124 -0.316938966 -1.022683457 -1.148827949
351 SOLV HW1 1125 -0.277201269 -1.112909811 -1.132088457
351 SOLV HW2 1126 -0.412561815 -1.022443252 -1.119533127
352 SOLV OW 1127 -1.526225970 0.310753091 -0.118555217
352 SOLV HW1 1128 -1.519469852 0.219998677 -0.077105472
352 SOLV HW2 1129 -1.435689292 0.340215446 -0.149168089
353 SOLV OW 1130 -0.391499023 1.210067984 1.234055044
353 SOLV HW1 1131 -0.347250104 1.199912821 1.323164301
353 SOLV HW2 1132 -0.401805258 1.120139195 1.191533349
354 SOLV OW 1133 0.964502961 1.355331208 -1.003928460
354 SOLV HW1 1134 1.064447216 1.352019397 -1.005283206
354 SOLV HW2 1135 0.934509572 1.434354594 -0.950475105
355 SOLV OW 1136 -1.364805916 0.369185108 1.507127194
355 SOLV HW1 1137 -1.290827138 0.353310905 1.572512841
355 SOLV HW2 1138 -1.378323613 0.287147057 1.451565335
356 SOLV OW 1139 -1.359605101 0.750994263 1.309547363
356 SOLV HW1 1140 -1.278869902 0.692459810 1.302085816
356 SOLV HW2 1141 -1.438407394 0.695744333 1.336708622
357 SOLV OW 1142 -0.512154401 -0.960030211 1.079764864
357 SOLV HW1 1143 -0.444830103 -0.944137694 1.007540415
357 SOLV HW2 1144 -0.498188272 -1.050996639 1.118899633
358 SOLV OW 1145 -1.031587026 0.222151022 -1.374361424
358 SOLV HW1 1146 -0.970311204 0.249231608 -1.300108401
358 SOLV HW2 1147 -1.091337069 0.148338787 -1.343005747
359 SOLV OW 1148 0.254271653 0.977192163 1.414760701
359 SOLV HW1 1149 0.293178190 0.937205888 1.331761048
359 SOLV HW2 1150 0.317878636 0.964424820 1.490870512
360 SOLV OW 1151 -1.304273167 0.794595230 -0.812943735
360 SOLV HW1 1152 -1.341165626 0.885226734 -0.833595896
360 SOLV HW2 1153 -1.276790286 0.749649597 -0.897950067
361 SOLV OW 1154 0.174001603 1.231660190 -0.685444326
361 SOLV HW1 1155 0.233245967 1.221633101 -0.765391057
361 SOLV HW2 1156 0.172716711 1.145923496 -0.633971619
362 SOLV OW 1157 0.648478965 0.827225473 1.026964198
362 SOLV HW1 1158 0.628107039 0.886822740 0.949289989
362 SOLV HW2 1159 0.745595614 0.834236382 1.049752216
363 SOLV OW 1160 1.264892372 1.142628012 0.294910015
363 SOLV HW1 1161 1.300739411 1.112604308 0.383312894
363 SOLV HW2 1162 1.173804929 1.103770868 0.280958551
364 SOLV OW 1163 0.682913062 0.654090378 0.307633064
364 SOLV HW1 1164 0.637757363 0.718447399 0.245819855
364 SOLV HW2 1165 0.620732176 0.630399620 0.382291593
365 SOLV OW 1166 1.165106165 0.653432064 0.213285509
365 SOLV HW1 1167 1.130901486 0.594932850 0.139746330
365 SOLV HW2 1168 1.216530085 0.598014911 0.278742439
366 SOLV OW 1169 0.749263613 -0.201287911 0.637975204
366 SOLV HW1 1170 0.751748957 -0.182432062 0.736158723
366 SOLV HW2 1171 0.817932476 -0.145326154 0.591556865
367 SOLV OW 1172 -0.137969389 -1.241581362 1.109687382
367 SOLV HW1 1173 -0.215278078 -1.220492900 1.169522925
367 SOLV HW2 1174 -0.161627519 -1.318747545 1.050634548
368 SOLV OW 1175 -1.035897938 -0.841402384 -0.614208857
368 SOLV HW1 1176 -1.131473603 -0.824819071 -0.589947024
368 SOLV HW2 1177 -1.031096709 -0.880183370 -0.706248690
369 SOLV OW 1178 -0.105855862 0.419799871 -1.416602863
369 SOLV HW1 1179 -0.058780938 0.498748411 -1.377198545
369 SOLV HW2 1180 -0.046190574 0.375388375 -1.483455723
370 SOLV OW 1181 -1.498472277 0.025246105 0.958278666
370 SOLV HW1 1182 -1.487205684 0.115872500 0.999039790
370 SOLV HW2 1183 -1.418225559 0.003505173 0.902695978
371 SOLV OW 1184 -0.665235187 -0.749811339 1.201050740
371 SOLV HW1 1185 -0.638958907 -0.666964468 1.151578548
371 SOLV HW2 1186 -0.618365546 -0.828672729 1.161230257
372 SOLV OW 1187 1.438179794 0.099715114 -0.515301996
372 SOLV HW1 1188 1.338915291 0.098862621 -0.503156663
372 SOLV HW2 1189 1.463869879 0.176284035 -0.574283870
373 SOLV OW 1190 -0.937424264 1.053533579 1.410376352
373 SOLV HW1 1191 -0.963426236 1.098203178 1.324758003
373 SOLV HW2 1192 -0.986223349 0.966642057 1.418772720
374 SOLV OW 1193 -0.190943771 -0.793665881 -1.260867914
374 SOLV HW1 1194 -0.211928493 -0.884519815 -1.224715246
374 SOLV HW2 1195 -0.100043073 -0.765687590 -1.229945842
375 SOLV OW 1196 1.363021051 -1.255612166 -0.453563266
375 SOLV HW1 1197 1.279533977 -1.277235126 -0.504183875
375 SOLV HW2 1198 1.412500752 -1.340112471 -0.433275543
376 SOLV OW 1199 -0.776120237 -1.144589623 -1.491652128
376 SOLV HW1 1200 -0.706332166 -1.086607896 -1.449583884
376 SOLV HW2 1201 -0.830164705 -1.090331095 -1.555972061
377 SOLV OW 1202 0.388306571 0.195425484 -0.432703801
377 SOLV HW1 1203 0.325601906 0.270732834 -0.412737683
377 SOLV HW2 1204 0.479083874 0.217269436 -0.396870918
378 SOLV OW 1205 0.351953703 1.351751822 1.222190296
378 SOLV HW1 1206 0.422075475 1.388930125 1.283024827
378 SOLV HW2 1207 0.281210546 1.306606737 1.276572435
379 SOLV OW 1208 1.275234727 -1.562836585 -0.697732244
379 SOLV HW1 1209 1.246315202 -1.475595627 -0.658308914
379 SOLV HW2 1210 1.303816731 -1.548719364 -0.792522538
380 SOLV OW 1211 0.818393535 -1.074803896 -1.355166184
380 SOLV HW1 1212 0.838949696 -1.016026866 -1.433414476
380 SOLV HW2 1213 0.760263422 -1.025228373 -1.290627638
381 SOLV OW 1214 0.499629184 1.491495750 1.422988624
381 SOLV HW1 1215 0.506590868 1.585419651 1.389352472
381 SOLV HW2 1216 0.556093192 1.481164748 1.504882723
382 SOLV OW 1217 -1.072977724 0.020963689 -0.793542762
382 SOLV HW1 1218 -1.025178727 0.073477469 -0.723131893
382 SOLV HW2 1219 -1.145390722 -0.033419168 -0.751128701
383 SOLV OW 1220 0.757443740 1.106792742 -1.249694791
383 SOLV HW1 1221 0.800523099 1.196942886 -1.254021691
383 SOLV HW2 1222 0.789930408 1.058204877 -1.168544422
384 SOLV OW 1223 -0.978580929 0.756190819 1.355288290
384 SOLV HW1 1224 -1.049281324 0.688829167 1.333707470
384 SOLV HW2 1225 -0.918317233 0.719664002 1.426251642
385 SOLV OW 1226 0.393444690 0.369274638 1.196718722
385 SOLV HW1 1227 0.457560171 0.429173080 1.148726625
385 SOLV HW2 1228 0.316185011 0.423211247 1.230239351
386 SOLV OW 1229 -1.315403055 0.856220160 -0.198144688
386 SOLV HW1 1230 -1.340891327 0.942456957 -0.154381090
386 SOLV HW2 1231 -1.221414289 0.862417767 -0.231748845
387 SOLV OW 1232 -0.707763335 0.571834755 -1.064453867
387 SOLV HW1 1233 -0.648861151 0.609760300 -1.135827045
387 SOLV HW2 1234 -0.784787225 0.633603922 -1.048520756
388 SOLV OW 1235 -0.765246868 0.095680496 -0.321843089
388 SOLV HW1 1236 -0.720772402 0.040827631 -0.392659266
388 SOLV HW2 1237 -0.773530474 0.041612973 -0.238118744
389 SOLV OW 1238 1.538824221 -0.049915378 -0.265045598
389 SOLV HW1 1239 1.466432480 -0.079384314 -0.202654549
389 SOLV HW2 1240 1.498413519 -0.022070746 -0.352184385
390 SOLV OW 1241 0.195536803 -1.425238995 0.380844543
390 SOLV HW1 1242 0.265757958 -1.491448580 0.354664964
390 SOLV HW2 1243 0.170249017 -1.439584492 0.476535605
391 SOLV OW 1244 1.300321983 0.671170916 -0.337771593
391 SOLV HW1 1245 1.279326679 0.768710457 -0.344638799
391 SOLV HW2 1246 1.371583234 0.657195291 -0.269008780
392 SOLV OW 1247 1.049435399 1.450117907 -0.067743136
392 SOLV HW1 1248 1.102044765 1.404001203 0.003721156
392 SOLV HW2 1249 1.002209353 1.529115969 -0.028620607
393 SOLV OW 1250 -0.164284323 -0.177359981 -0.239334083
393 SOLV HW1 1251 -0.198642967 -0.245068649 -0.174256050
393 SOLV HW2 1252 -0.064292771 -0.178670843 -0.239086497
394 SOLV OW 1253 -0.149310004 0.773086152 -1.304127218
394 SOLV HW1 1254 -0.114382815 0.856888326 -1.262182564
394 SOLV HW2 1255 -0.081778050 0.700040050 -1.293842501
395 SOLV OW 1256 1.453488104 0.727091048 -0.098488470
395 SOLV HW1 1257 1.408679008 0.642154924 -0.070568183
395 SOLV HW2 1258 1.445720578 0.794550942 -0.025068957
396 SOLV OW 1259 -1.170257556 -0.459264782 -0.036180230
396 SOLV HW1 1260 -1.263563280 -0.477845879 -0.005376417
396 SOLV HW2 1261 -1.168620345 -0.453734452 -0.136023884
397 SOLV OW 1262 1.222305554 -0.367746352 -1.122754902
397 SOLV HW1 1263 1.232356447 -0.461515391 -1.089491114
397 SOLV HW2 1264 1.157263661 -0.318640540 -1.064804393
398 SOLV OW 1265 -0.045421070 0.046336372 1.422760080
398 SOLV HW1 1266 -0.133393724 0.072262780 1.382900726
398 SOLV HW2 1267 -0.041493443 0.077271945 1.517774048
399 SOLV OW 1268 -0.411248857 1.263643557 -0.597685455
399 SOLV HW1 1269 -0.478550246 1.331548093 -0.568341460
399 SOLV HW2 1270 -0.342057839 1.252334322 -0.526367600
400 SOLV OW 1271 0.600275836 0.403285625 -1.411709842
400 SOLV HW1 1272 0.589385389 0.502615384 -1.415837407
400 SOLV HW2 1273 0.554529242 0.361469552 -1.490199408
401 SOLV OW 1274 0.315553060 -1.529148749 -0.083616524
401 SOLV HW1 1275 0.372176917 -1.523992799 -0.001342949
401 SOLV HW2 1276 0.220001613 -1.512247847 -0.059407885
402 SOLV OW 1277 -0.485763830 0.709624841 -1.220378967
402 SOLV HW1 1278 -0.482036235 0.740359261 -1.315466246
402 SOLV HW2 1279 -0.414584941 0.641145280 -1.204690698
403 SOLV OW 1280 -1.347267263 0.690918925 -1.321230110
403 SOLV HW1 1281 -1.310184484 0.605799887 -1.358397858
403 SOLV HW2 1282 -1.331551237 0.693797459 -1.222505646
404 SOLV OW 1283 0.246090289 1.009121205 0.459182456
404 SOLV HW1 1284 0.198516308 0.933239038 0.414681858
404 SOLV HW2 1285 0.248783988 1.087633446 0.397294195
405 SOLV OW 1286 0.840481921 0.613094733 -0.478036706
405 SOLV HW1 1287 0.809580084 0.618350226 -0.383075724
405 SOLV HW2 1288 0.764510663 0.584071036 -0.536226767
406 SOLV OW 1289 -0.252863340 -1.418416141 -0.179645369
406 SOLV HW1 1290 -0.282774537 -1.462437273 -0.094974534
406 SOLV HW2 1291 -0.198233164 -1.482837978 -0.233189480
407 SOLV OW 1292 0.035006646 -0.633859211 -0.334061865
407 SOLV HW1 1293 -0.013534383 -0.590567382 -0.410030849
407 SOLV HW2 1294 0.066722850 -0.563549705 -0.270404113
408 SOLV OW 1295 -1.325183206 0.921240197 0.740603059
408 SOLV HW1 1296 -1.295526400 0.861412433 0.666163506
408 SOLV HW2 1297 -1.267587326 0.905621169 0.820845509
409 SOLV OW 1298 -1.360034205 -0.268983871 -0.990036832
409 SOLV HW1 1299 -1.382214469 -0.260120907 -0.892922762
409 SOLV HW2 1300 -1.377564436 -0.362842648 -1.019782625
410 SOLV OW 1301 0.178477027 1.341119908 0.856870845
410 SOLV HW1 1302 0.101112964 1.277852030 0.860610230
410 SOLV HW2 1303 0.241346444 1.312621407 0.784501981
411 SOLV OW 1304 0.780298143 -1.319060687 -1.477260211
411 SOLV HW1 1305 0.835348192 -1.376501398 -1.416664628
411 SOLV HW2 1306 0.771841513 -1.227550344 -1.437813837
412 SOLV OW 1307 -0.574302381 -0.810941940 1.457718343
412 SOLV HW1 1308 -0.550580029 -0.902920325 1.489003475
412 SOLV HW2 1309 -0.579728848 -0.810081802 1.357861724
413 SOLV OW 1310 1.155802355 0.178907875 -0.742335843
413 SOLV HW1 1311 1.238664458 0.226353795 -0.772080907
413 SOLV HW2 1312 1.120326764 0.222508048 -0.659616671
414 SOLV OW 1313 -0.085479861 1.308899267 0.667177630
414 SOLV HW1 1314 -0.103728162 1.370173108 0.744083596
414 SOLV HW2 1315 -0.021277361 1.352671601 0.604218065
415 SOLV OW 1316 0.947644966 -0.381648829 -0.511409995
415 SOLV HW1 1317 0.973829944 -0.399017544 -0.606355179
415 SOLV HW2 1318 0.878872530 -0.448495267 -0.483059403
416 SOLV OW 1319 -1.210456284 -0.681162341 1.326782209
416 SOLV HW1 1320 -1.117174903 -0.646163782 1.335464547
416 SOLV HW2 1321 -1.209317409 -0.780963606 1.333104519
417 SOLV OW 1322 -1.198604350 -0.415364370 -0.301735136
417 SOLV HW1 1323 -1.142192099 -0.493928297 -0.327140990
417 SOLV HW2 1324 -1.175854741 -0.337650068 -0.360429498
418 SOLV OW 1325 0.948086406 0.353029575 -0.227084876
418 SOLV HW1 1326 0.925587211 0.445096005 -0.195154482
418 SOLV HW2 1327 0.864355955 0.298774429 -0.233981453
419 SOLV OW 1328 0.130734373 -1.272422161 0.136962381
419 SOLV HW1 1329 0.151227889 -1.306903183 0.228565910
419 SOLV HW2 1330 0.152007983 -1.174808782 0.132586114
420 SOLV OW 1331 -1.173768099 1.549799142 -0.331684696
420 SOLV HW1 1332 -1.175314022 1.451584477 -0.350478613
420 SOLV HW2 1333 -1.237277310 1.596497970 -0.393227114
421 SOLV OW 1334 -1.296579346 -1.240873742 0.548815789
421 SOLV HW1 1335 -1.345316510 -1.249241444 0.635745049
421 SOLV HW2 1336 -1.351214933 -1.282096997 0.475893476
422 SOLV OW 1337 -0.018062993 1.241054182 0.113569509
422 SOLV HW1 1338 0.063861620 1.261173807 0.059851974
422 SOLV HW2 1339 -0.096135478 1.229415800 0.052160313
423 SOLV OW 1340 -1.122934393 -0.561199480 0.793027042
423 SOLV HW1 1341 -1.106660192 -0.634596814 0.858979411
423 SOLV HW2 1342 -1.197811301 -0.503335531 0.825382463
424 SOLV OW 1343 -1.396034253 0.659156389 -0.015491803
424 SOLV HW1 1344 -1.360959567 0.712360166 -0.092570879
424 SOLV HW2 1345 -1.319457796 0.620021390 0.035561699
425 SOLV OW 1346 1.309322564 -0.248632837 0.839433450
425 SOLV HW1 1347 1.385421352 -0.201589171 0.884129421
425 SOLV HW2 1348 1.232817746 -0.256441347 0.903369541
426 SOLV OW 1349 0.601069234 -1.535608130 -1.519037433
426 SOLV HW1 1350 0.540797596 -1.508581201 -1.594128051
426 SOLV HW2 1351 0.660068110 -1.458723269 -1.494350625
427 SOLV OW 1352 -0.945869294 0.167753816 -0.596141703
427 SOLV HW1 1353 -0.978356227 0.248873539 -0.644782684
427 SOLV HW2 1354 -0.872526365 0.193729823 -0.533310648
428 SOLV OW 1355 0.544592038 -1.484805095 0.106100909
428 SOLV HW1 1356 0.534746420 -1.495762633 0.205010599
428 SOLV HW2 1357 0.589358985 -1.397548222 0.086549724
429 SOLV OW 1358 1.305568036 0.973870758 1.473595231
429 SOLV HW1 1359 1.224417571 0.934680903 1.430228452
429 SOLV HW2 1360 1.387531219 0.930139811 1.436562511
430 SOLV OW 1361 -1.332560263 -0.407537331 0.891975117
430 SOLV HW1 1362 -1.396359942 -0.390149110 0.816950322
430 SOLV HW2 1363 -1.380557339 -0.453911197 0.966454429
431 SOLV OW 1364 0.258484406 -0.614710806 -0.527307004
431 SOLV HW1 1365 0.218922258 -0.642260771 -0.439684918
431 SOLV HW2 1366 0.274908315 -0.695845349 -0.583424468
432 SOLV OW 1367 -1.164324780 0.569707339 1.230991132
432 SOLV HW1 1368 -1.157420960 0.614788583 1.141987155
432 SOLV HW2 1369 -1.106888723 0.487838063 1.231412332
433 SOLV OW 1370 0.679393891 1.127131508 0.043164076
433 SOLV HW1 1371 0.711508245 1.118437994 0.137476246
433 SOLV HW2 1372 0.644926312 1.038776977 0.011429539
434 SOLV OW 1373 1.033951060 -0.753088172 -1.485403999
434 SOLV HW1 1374 1.085900338 -0.821499044 -1.434186951
434 SOLV HW2 1375 1.097083859 -0.687801187 -1.527281611
435 SOLV OW 1376 -1.137525503 1.431666757 -1.288496739
435 SOLV HW1 1377 -1.088229904 1.453722835 -1.204332594
435 SOLV HW2 1378 -1.095886856 1.480425100 -1.365236150
436 SOLV OW 1379 -0.826703553 0.729366990 0.923027541
436 SOLV HW1 1380 -0.735608143 0.690542589 0.936971239
436 SOLV HW2 1381 -0.832033491 0.769867394 0.831751043
437 SOLV OW 1382 0.704737512 -0.757329735 0.944655482
437 SOLV HW1 1383 0.699735610 -0.853797135 0.918752930
437 SOLV HW2 1384 0.786015840 -0.742113603 1.000907140
438 SOLV OW 1385 -1.058835467 0.800760560 0.054962076
438 SOLV HW1 1386 -1.133320666 0.859817741 0.086037539
438 SOLV HW2 1387 -1.086488259 0.705023692 0.063402792
439 SOLV OW 1388 -0.869259318 -0.972103565 0.883447535
439 SOLV HW1 1389 -0.781035482 -1.019155148 0.881390561
439 SOLV HW2 1390 -0.854985751 -0.873962601 0.870560704
440 SOLV OW 1391 -1.429754578 -1.234601297 0.786431613
440 SOLV HW1 1392 -1.525558930 -1.239605895 0.758181835
440 SOLV HW2 1393 -1.418703094 -1.279875863 0.874916422
441 SOLV OW 1394 -0.315194744 -0.687185827 -0.082601049
441 SOLV HW1 1395 -0.305156667 -0.755446275 -0.154999662
441 SOLV HW2 1396 -0.407866288 -0.649651132 -0.084821850
442 SOLV OW 1397 -0.836643868 -0.417189822 -0.535729636
442 SOLV HW1 1398 -0.769303329 -0.384304527 -0.601955344
442 SOLV HW2 1399 -0.841078969 -0.517030113 -0.539513528
443 SOLV OW 1400 1.318179096 0.689656732 1.198017576
443 SOLV HW1 1401 1.336531451 0.642992886 1.284548626
443 SOLV HW2 1402 1.368545901 0.644390876 1.124423910
444 SOLV OW 1403 1.403025431 -0.804653030 -0.348805116
444 SOLV HW1 1404 1.427959496 -0.823414454 -0.253787928
444 SOLV HW2 1405 1.438198799 -0.714881527 -0.375371498
445 SOLV OW 1406 -0.446686691 0.713491511 1.189524946
445 SOLV HW1 1407 -0.352293914 0.740759049 1.208186486
445 SOLV HW2 1408 -0.504370649 0.794979585 1.183699675
446 SOLV OW 1409 -1.240532580 -0.264058562 0.436328464
446 SOLV HW1 1410 -1.198366151 -0.347345960 0.472204767
446 SOLV HW2 1411 -1.185475430 -0.184834944 0.462675708
447 SOLV OW 1412 -0.341491488 1.216997646 0.546997221
447 SOLV HW1 1413 -0.393284740 1.143114792 0.590128467
447 SOLV HW2 1414 -0.262078525 1.239410646 0.603501633
448 SOLV OW 1415 -1.091761733 0.587387405 -0.202199194
448 SOLV HW1 1416 -0.994807292 0.562885685 -0.201198740
448 SOLV HW2 1417 -1.107968543 0.656362470 -0.272776850
449 SOLV OW 1418 0.097397447 -1.492125454 0.615175561
449 SOLV HW1 1419 0.025893532 -1.562042926 0.615922890
449 SOLV HW2 1420 0.080000798 -1.425904390 0.688072086
450 SOLV OW 1421 -0.746740468 0.936539676 -1.304647706
450 SOLV HW1 1422 -0.676985141 0.906755485 -1.369818451
450 SOLV HW2 1423 -0.704992600 0.949271280 -1.214674866
451 SOLV OW 1424 -1.054752232 -1.457679649 -1.302595906
451 SOLV HW1 1425 -1.044706112 -1.525140972 -1.375738939
451 SOLV HW2 1426 -0.991732194 -1.381670231 -1.318496347
452 SOLV OW 1427 -1.330810412 0.335858146 1.039987145
452 SOLV HW1 1428 -1.274173509 0.348580633 0.958548144
452 SOLV HW2 1429 -1.272202063 0.315991373 1.118550823
453 SOLV OW 1430 0.658677820 -1.053902552 1.497415758
453 SOLV HW1 1431 0.749699629 -1.079047827 1.464482275
453 SOLV HW2 1432 0.632074915 -0.965963781 1.457907395
454 SOLV OW 1433 0.531040824 -0.640087262 -0.121089172
454 SOLV HW1 1434 0.459507215 -0.706214967 -0.098459788
454 SOLV HW2 1435 0.598259056 -0.683059527 -0.181396849
455 SOLV OW 1436 -0.928197102 1.243291333 0.080968260
455 SOLV HW1 1437 -0.990505199 1.220255510 0.155728819
455 SOLV HW2 1438 -0.836282382 1.257990167 0.117542140
456 SOLV OW 1439 -1.490538181 0.067278387 0.051165439
456 SOLV HW1 1440 -1.486402348 -0.023788826 0.010039727
456 SOLV HW2 1441 -1.410751899 0.080968713 0.109887968
457 SOLV OW 1442 -0.185913382 -1.103899832 0.001114596
457 SOLV HW1 1443 -0.279211440 -1.101009802 -0.034742403
457 SOLV HW2 1444 -0.186934375 -1.141558501 0.093739308
458 SOLV OW 1445 -0.851935708 -0.140326284 -0.740897601
458 SOLV HW1 1446 -0.863333854 -0.225548234 -0.791974219
458 SOLV HW2 1447 -0.929179887 -0.079644850 -0.759673970
459 SOLV OW 1448 1.005827401 0.375350811 1.243492709
459 SOLV HW1 1449 1.022823465 0.473562460 1.235296245
459 SOLV HW2 1450 0.907248372 0.358693362 1.245986986
460 SOLV OW 1451 -0.556392323 0.665007090 0.948091462
460 SOLV HW1 1452 -0.513384546 0.685841784 1.035943578
460 SOLV HW2 1453 -0.518050549 0.724363655 0.877321077
461 SOLV OW 1454 1.290505700 -0.540538019 -0.752004407
461 SOLV HW1 1455 1.211511833 -0.480432711 -0.739861420
461 SOLV HW2 1456 1.279376373 -0.622060533 -0.695168609
462 SOLV OW 1457 0.665729565 0.594755814 1.304020982
462 SOLV HW1 1458 0.628847233 0.602876583 1.211416196
462 SOLV HW2 1459 0.763761997 0.614496719 1.302683841
463 SOLV OW 1460 0.479915639 -0.034897990 0.789030514
463 SOLV HW1 1461 0.425561488 0.017680580 0.854472620
463 SOLV HW2 1462 0.575475538 -0.036155135 0.818496275
464 SOLV OW 1463 1.189861420 -1.479841456 -0.306445491
464 SOLV HW1 1464 1.283378820 -1.496767831 -0.337560189
464 SOLV HW2 1465 1.127922182 -1.480972655 -0.384946073
465 SOLV OW 1466 -1.074559732 0.831341810 -0.332357078
465 SOLV HW1 1467 -0.978710726 0.857654917 -0.343427133
465 SOLV HW2 1468 -1.123117613 0.845301490 -0.418665740
466 SOLV OW 1469 0.217556957 -0.435498731 -1.049247947
466 SOLV HW1 1470 0.221059446 -0.371486494 -0.972499942
466 SOLV HW2 1471 0.277117258 -0.513563061 -1.030258374
467 SOLV OW 1472 0.305170104 1.289192396 -1.130284733
467 SOLV HW1 1473 0.329563401 1.242961575 -1.215545240
467 SOLV HW2 1474 0.206008142 1.301535936 -1.126270687
468 SOLV OW 1475 0.443201706 1.482623836 0.598645831
468 SOLV HW1 1476 0.485186182 1.441575518 0.679593175
468 SOLV HW2 1477 0.503935496 1.552929799 0.561651662
469 SOLV OW 1478 0.816515544 0.735574331 0.745965157
469 SOLV HW1 1479 0.866213605 0.761762197 0.828707974
469 SOLV HW2 1480 0.871863554 0.671498042 0.692740093
470 SOLV OW 1481 -1.156875489 -1.052986990 0.861079188
470 SOLV HW1 1482 -1.183088181 -1.143992128 0.893219312
470 SOLV HW2 1483 -1.064874786 -1.031332952 0.893773688
471 SOLV OW 1484 -0.790811469 -0.764619680 -0.943501743
471 SOLV HW1 1485 -0.839024084 -0.842581216 -0.903531343
471 SOLV HW2 1486 -0.856519941 -0.704755830 -0.989335591
472 SOLV OW 1487 0.038215939 -0.953846156 -1.469404741
472 SOLV HW1 1488 0.059556387 -0.876977661 -1.409090709
472 SOLV HW2 1489 -0.041738664 -1.002567822 -1.434254733
473 SOLV OW 1490 0.002802818 1.331006653 -1.099776524
473 SOLV HW1 1491 -0.041479205 1.420608460 -1.103283052
473 SOLV HW2 1492 0.016169320 1.296788396 -1.192793070
474 SOLV OW 1493 -1.193680202 0.475363862 -1.436360225
474 SOLV HW1 1494 -1.098959641 0.504761006 -1.423555750
474 SOLV HW2 1495 -1.204909773 0.382661283 -1.400580082
475 SOLV OW 1496 1.243157080 0.018089028 -1.030361543
475 SOLV HW1 1497 1.326334627 -0.033087448 -1.008821403
475 SOLV HW2 1498 1.263749880 0.115938150 -1.032087782
476 SOLV OW 1499 0.728606872 0.321880324 1.182120529
476 SOLV HW1 1500 0.725648337 0.269706301 1.096860621
476 SOLV HW2 1501 0.678615880 0.407692917 1.170407721
477 SOLV OW 1502 -0.695485298 -0.042210887 0.335507724
477 SOLV HW1 1503 -0.731993205 0.049913547 0.348996852
477 SOLV HW2 1504 -0.652725774 -0.073832388 0.420203748
478 SOLV OW 1505 -1.069496577 -1.389675599 0.217891965
478 SOLV HW1 1506 -1.027125323 -1.371616071 0.129119550
478 SOLV HW2 1507 -1.034043770 -1.325054861 0.285487682
479 SOLV OW 1508 -1.273873001 -0.726109242 0.189653144
479 SOLV HW1 1509 -1.321506578 -0.639734895 0.173161366
479 SOLV HW2 1510 -1.322854462 -0.800408856 0.144026072
480 SOLV OW 1511 -0.814285812 1.413278648 -1.283132915
480 SOLV HW1 1512 -0.727421524 1.456922687 -1.306583357
480 SOLV HW2 1513 -0.879216308 1.425897766 -1.358132106
481 SOLV OW 1514 1.381284325 0.318301697 -0.808524495
481 SOLV HW1 1515 1.452301200 0.382878737 -0.780455201
481 SOLV HW2 1516 1.371234271 0.320991136 -0.907989047
482 SOLV OW 1517 -1.548315847 0.594739568 -1.000637636
482 SOLV HW1 1518 -1.474805407 0.602469778 -0.933270724
482 SOLV HW2 1519 -1.572068590 0.685750948 -1.034616291
483 SOLV OW 1520 -0.531548281 0.940709856 -1.123294467
483 SOLV HW1 1521 -0.505086601 0.847812792 -1.149213274
483 SOLV HW2 1522 -0.463047061 0.978745815 -1.061144980
484 SOLV OW 1523 0.219503171 0.901050088 1.109150594
484 SOLV HW1 1524 0.306927496 0.947832802 1.122197762
484 SOLV HW2 1525 0.176471749 0.933720562 1.024992070
485 SOLV OW 1526 0.712925587 0.481594895 0.788883793
485 SOLV HW1 1527 0.730176778 0.580051206 0.785686860
485 SOLV HW2 1528 0.744015659 0.439537699 0.703642703
486 SOLV OW 1529 -1.521295712 1.124582993 -0.231919981
486 SOLV HW1 1530 -1.572139515 1.082515749 -0.307068298
486 SOLV HW2 1531 -1.517526187 1.061195454 -0.154655590
487 SOLV OW 1532 1.025500417 -0.536631254 0.257139315
487 SOLV HW1 1533 1.121936003 -0.544041473 0.282570820
487 SOLV HW2 1534 0.968536048 -0.568970397 0.332708692
488 SOLV OW 1535 0.222006785 0.625022902 1.072123486
488 SOLV HW1 1536 0.214666014 0.723165712 1.089895153
488 SOLV HW2 1537 0.204042746 0.574450497 1.156511480
489 SOLV OW 1538 1.485332377 1.065670939 -1.174276126
489 SOLV HW1 1539 1.465473216 1.131167049 -1.247198434
489 SOLV HW2 1540 1.584276110 1.053237947 -1.166701655
490 SOLV OW 1541 1.192255082 -1.000963256 0.210627461
490 SOLV HW1 1542 1.250753796 -0.929331580 0.248690070
490 SOLV HW2 1543 1.124873268 -0.959821855 0.149235357
491 SOLV OW 1544 0.476298819 -0.914474926 1.292849588
491 SOLV HW1 1545 0.428901740 -0.848965099 1.233998716
491 SOLV HW2 1546 0.427413036 -1.001712349 1.291729688
492 SOLV OW 1547 0.892505470 -0.303652211 1.151337603
492 SOLV HW1 1548 0.815661235 -0.356885365 1.115800159
492 SOLV HW2 1549 0.857841665 -0.228577170 1.207585900
493 SOLV OW 1550 -0.095359353 -0.521868893 1.202862982
493 SOLV HW1 1551 -0.167531476 -0.586774389 1.178778796
493 SOLV HW2 1552 -0.136573668 -0.435695486 1.232477800
494 SOLV OW 1553 -0.154214700 -1.251420894 -0.506647196
494 SOLV HW1 1554 -0.066322132 -1.271000166 -0.463133204
494 SOLV HW2 1555 -0.210984582 -1.333748860 -0.505585306
495 SOLV OW 1556 -0.500697196 0.839704735 -0.225335811
495 SOLV HW1 1557 -0.493783374 0.811844372 -0.321137655
495 SOLV HW2 1558 -0.530163743 0.935150131 -0.220445239
496 SOLV OW 1559 1.526914062 0.818623463 0.379258366
496 SOLV HW1 1560 1.450405286 0.811426566 0.443233990
496 SOLV HW2 1561 1.541139890 0.914562441 0.354933215
497 SOLV OW 1562 0.343002126 -0.838704637 -1.233080031
497 SOLV HW1 1563 0.382544090 -0.888035538 -1.155591064
497 SOLV HW2 1564 0.412113484 -0.828082196 -1.304581169
498 SOLV OW 1565 0.780763674 -0.976711841 0.793048504
498 SOLV HW1 1566 0.727670297 -1.060516680 0.805678182
498 SOLV HW2 1567 0.877301621 -0.999920619 0.781073547
499 SOLV OW 1568 -0.335226509 -0.471079001 -0.441674390
499 SOLV HW1 1569 -0.238118266 -0.447990540 -0.447760629
499 SOLV HW2 1570 -0.346322583 -0.570274546 -0.447774490
500 SOLV OW 1571 -0.572809466 -0.874839754 -1.097599771
500 SOLV HW1 1572 -0.518382607 -0.799225173 -1.133956574
500 SOLV HW2 1573 -0.647889021 -0.838372439 -1.042510502
501 SOLV OW 1574 0.627366464 0.875682383 -0.045207808
501 SOLV HW1 1575 0.650303251 0.919006701 -0.132378819
501 SOLV HW2 1576 0.594921468 0.782551079 -0.061813068
502 SOLV OW 1577 -1.553869452 -1.207937627 -0.111251763
502 SOLV HW1 1578 -1.477789896 -1.272404092 -0.103668275
502 SOLV HW2 1579 -1.571380469 -1.188268657 -0.207729973
503 SOLV OW 1580 1.367896197 1.514933533 1.291414646
503 SOLV HW1 1581 1.360300311 1.581523860 1.217186124
503 SOLV HW2 1582 1.321047055 1.430507328 1.265352430
504 SOLV OW 1583 -0.363947052 0.075585237 -1.421184614
504 SOLV HW1 1584 -0.324762149 0.138620998 -1.354157174
504 SOLV HW2 1585 -0.308054212 -0.007223769 -1.425690051
505 SOLV OW 1586 1.309675658 -1.002274059 0.873622173
505 SOLV HW1 1587 1.224653638 -1.040390091 0.909956487
505 SOLV HW2 1588 1.362540491 -0.961701066 0.948193339
506 SOLV OW 1589 -1.197258001 0.896141350 -0.564551267
506 SOLV HW1 1590 -1.203418581 0.833740586 -0.642459583
506 SOLV HW2 1591 -1.191724074 0.990342340 -0.597671944
507 SOLV OW 1592 -0.428932517 1.353361955 0.123053533
507 SOLV HW1 1593 -0.366630111 1.299270656 0.179569854
507 SOLV HW2 1594 -0.380098131 1.432287195 0.085804099
508 SOLV OW 1595 -0.634980940 1.413213699 0.783370137
508 SOLV HW1 1596 -0.551491950 1.464663041 0.802896812
508 SOLV HW2 1597 -0.612271321 1.331860473 0.729847858
509 SOLV OW 1598 0.229788566 -0.611487764 0.927964303
509 SOLV HW1 1599 0.283963002 -0.536929389 0.966794837
509 SOLV HW2 1600 0.242512711 -0.613773121 0.828795859
510 SOLV OW 1601 0.148897488 0.038836285 -1.319288614
510 SOLV HW1 1602 0.151896053 -0.023401186 -1.397515222
510 SOLV HW2 1603 0.053703150 0.060456701 -1.297551165
511 SOLV OW 1604 1.213777687 -0.087321453 -0.726659361
511 SOLV HW1 1605 1.214189195 0.004510552 -0.687053269
511 SOLV HW2 1606 1.224099237 -0.080834925 -0.825922793
512 SOLV OW 1607 0.440185396 -1.367978306 -0.585375948
512 SOLV HW1 1608 0.440393071 -1.341966249 -0.488808907
512 SOLV HW2 1609 0.346239160 -1.385185669 -0.615038565
513 SOLV OW 1610 -1.037128683 -0.133993991 1.280605747
513 SOLV HW1 1611 -0.986878233 -0.108974004 1.197837929
513 SOLV HW2 1612 -1.077306807 -0.224838512 1.269005987
514 SOLV OW 1613 0.481000329 -1.196239141 -1.303675201
514 SOLV HW1 1614 0.390151535 -1.165405946 -1.331916329
514 SOLV HW2 1615 0.479770033 -1.295284432 -1.289881732
515 SOLV OW 1616 0.166562157 -0.388356329 1.231031646
515 SOLV HW1 1617 0.159536032 -0.303894764 1.284125335
515 SOLV HW2 1618 0.075013980 -0.425174244 1.214741515
516 SOLV OW 1619 0.550240010 -1.264108495 0.640648103
516 SOLV HW1 1620 0.489940097 -1.221043315 0.707813590
516 SOLV HW2 1621 0.565898953 -1.201014503 0.564649688
517 SOLV OW 1622 1.199619887 0.928007889 -0.410485812
517 SOLV HW1 1623 1.159893807 1.013617620 -0.377405612
517 SOLV HW2 1624 1.181428239 0.918465798 -0.508360577
518 SOLV OW 1625 1.334067117 -1.480109214 0.935680853
518 SOLV HW1 1626 1.332457332 -1.439331489 1.026985695
518 SOLV HW2 1627 1.376972853 -1.416494668 0.871539073
519 SOLV OW 1628 1.123412112 0.330668825 -0.014270291
519 SOLV HW1 1629 1.088503202 0.263055482 0.050626913
519 SOLV HW2 1630 1.063707571 0.333819949 -0.094439867
520 SOLV OW 1631 -1.083053208 -0.646428231 -1.167994348
520 SOLV HW1 1632 -1.164842380 -0.589700956 -1.158367414
520 SOLV HW2 1633 -1.006169546 -0.589386597 -1.196894946
521 SOLV OW 1634 -1.011986776 -0.635609884 -0.266748964
521 SOLV HW1 1635 -0.940293941 -0.650091774 -0.334943834
521 SOLV HW2 1636 -0.972493618 -0.592741452 -0.185492099
522 SOLV OW 1637 0.782700829 -1.433997211 -1.140679148
522 SOLV HW1 1638 0.807204865 -1.369883246 -1.067953070
522 SOLV HW2 1639 0.865854965 -1.473933494 -1.179287088
523 SOLV OW 1640 1.474865083 -0.672853053 0.948700821
523 SOLV HW1 1641 1.526798885 -0.616046760 0.884856800
523 SOLV HW2 1642 1.397802202 -0.620382139 0.984869946
524 SOLV OW 1643 0.939644048 1.411874450 0.949714412
524 SOLV HW1 1644 0.880737134 1.357844627 0.889611718
524 SOLV HW2 1645 0.883072680 1.467890846 1.010239984
525 SOLV OW 1646 0.972377408 0.266455790 0.721046697
525 SOLV HW1 1647 1.037990585 0.295653354 0.651447223
525 SOLV HW2 1648 0.890079828 0.231152278 0.676523571
526 SOLV OW 1649 -0.364920323 0.070645618 -0.958614435
526 SOLV HW1 1650 -0.318182485 0.107446054 -0.878221898
526 SOLV HW2 1651 -0.309895560 0.088458502 -1.040202155
527 SOLV OW 1652 -0.309976038 1.326261624 -1.343351439
527 SOLV HW1 1653 -0.278886327 1.231376122 -1.348978852
527 SOLV HW2 1654 -0.408507710 1.328083444 -1.326333882
528 SOLV OW 1655 0.539098897 1.540578973 -0.488241209
528 SOLV HW1 1656 0.637401240 1.528980659 -0.502521928
528 SOLV HW2 1657 0.498878787 1.453208553 -0.460845534
529 SOLV OW 1658 -0.831493192 -0.423821674 1.101507293
529 SOLV HW1 1659 -0.797269354 -0.353555582 1.039113045
529 SOLV HW2 1660 -0.920192520 -0.456780021 1.069136605
530 SOLV OW 1661 0.311788933 0.792578062 -1.077477964
530 SOLV HW1 1662 0.300429089 0.699192260 -1.043535424
530 SOLV HW2 1663 0.394676332 0.832443305 -1.038204356
531 SOLV OW 1664 0.019095496 1.336975158 -1.369341276
531 SOLV HW1 1665 -0.079932678 1.334957734 -1.383106434
531 SOLV HW2 1666 0.065124416 1.314425674 -1.455207157
532 SOLV OW 1667 -1.035682809 0.992064461 0.472443503
532 SOLV HW1 1668 -1.082111831 1.072877308 0.436197678
532 SOLV HW2 1669 -0.965553494 0.962123641 0.407747935
533 SOLV OW 1670 0.918392377 1.317266435 -1.270967039
533 SOLV HW1 1671 0.939107087 1.322027830 -1.173241832
533 SOLV HW2 1672 0.958610162 1.396172225 -1.317425891
534 SOLV OW 1673 0.784130533 0.966673629 -0.568910964
534 SOLV HW1 1674 0.825949147 0.879577051 -0.594736582
534 SOLV HW2 1675 0.855238161 1.036430943 -0.560011749
535 SOLV OW 1676 -1.551758510 0.053491678 -1.319058547
535 SOLV HW1 1677 -1.507363842 0.003565181 -1.393477931
535 SOLV HW2 1678 -1.551250075 -0.002669582 -1.236309454
536 SOLV OW 1679 -1.553182779 0.554022304 1.478006344
536 SOLV HW1 1680 -1.495345484 0.476668456 1.503950558
536 SOLV HW2 1681 -1.535211669 0.631053360 1.539203822
537 SOLV OW 1682 1.026620198 1.027030009 0.832372499
537 SOLV HW1 1683 1.059875000 0.934938704 0.852739440
537 SOLV HW2 1684 1.057345722 1.090073340 0.903667343
538 SOLV OW 1685 0.119438201 -0.582727028 -0.800883701
538 SOLV HW1 1686 0.150703677 -0.579517536 -0.705942298
538 SOLV HW2 1687 0.194810489 -0.611848412 -0.859812722
539 SOLV OW 1688 -1.442592971 -0.334170191 -0.235521597
539 SOLV HW1 1689 -1.498975600 -0.363179960 -0.312861713
539 SOLV HW2 1690 -1.347829244 -0.362110453 -0.251052012
540 SOLV OW 1691 0.355423184 0.398979844 0.800981033
540 SOLV HW1 1692 0.436488565 0.446605822 0.766917864
540 SOLV HW2 1693 0.341790322 0.314843958 0.748680097
541 SOLV OW 1694 -0.049151947 -0.420986428 -0.550018449
541 SOLV HW1 1695 -0.086650446 -0.375559362 -0.630838606
541 SOLV HW2 1696 0.031092723 -0.370750545 -0.517788948
542 SOLV OW 1697 1.449257540 -0.203695920 0.125077406
542 SOLV HW1 1698 1.365529284 -0.152615289 0.105571473
542 SOLV HW2 1699 1.490525065 -0.234051522 0.039195983
543 SOLV OW 1700 1.445055586 -1.507316830 -0.376682041
543 SOLV HW1 1701 1.435950893 -1.574905148 -0.449828587
543 SOLV HW2 1702 1.541989374 -1.490155184 -0.359053657
544 SOLV OW 1703 1.112424992 -1.316388855 0.688243993
544 SOLV HW1 1704 1.106134851 -1.326973936 0.588996724
544 SOLV HW2 1705 1.079161752 -1.399520168 0.732789283
545 SOLV OW 1706 1.130115375 1.168216978 -0.835679669
545 SOLV HW1 1707 1.201467013 1.235433541 -0.855495864
545 SOLV HW2 1708 1.171789514 1.085596777 -0.797746456
546 SOLV OW 1709 0.913184208 1.037503956 1.419135899
546 SOLV HW1 1710 0.928939151 0.946703902 1.457977164
546 SOLV HW2 1711 0.820185363 1.042475060 1.382695673
547 SOLV OW 1712 -1.516265612 -0.511060988 -1.200281383
547 SOLV HW1 1713 -1.440565395 -0.529027603 -1.137442638
547 SOLV HW2 1714 -1.489273135 -0.536630052 -1.293123603
548 SOLV OW 1715 -0.824605836 0.914983429 -0.311294017
548 SOLV HW1 1716 -0.769518501 0.958326589 -0.382626377
548 SOLV HW2 1717 -0.797414496 0.950657542 -0.221909847
549 SOLV OW 1718 0.082516172 -0.722443155 -1.307740447
549 SOLV HW1 1719 0.045543801 -0.640003671 -1.264862981
549 SOLV HW2 1720 0.158009294 -0.757751367 -1.252461495
550 SOLV OW 1721 -0.905071334 -0.677909208 -1.460540590
550 SOLV HW1 1722 -0.933786672 -0.615812217 -1.533485522
550 SOLV HW2 1723 -0.817984490 -0.720383809 -1.485304192
551 SOLV OW 1724 -1.534322888 -1.250817194 -0.387793805
551 SOLV HW1 1725 -1.498395326 -1.344056467 -0.383603153
551 SOLV HW2 1726 -1.489939051 -1.201343390 -0.462522562
552 SOLV OW 1727 0.819294760 0.597534550 -1.096241992
552 SOLV HW1 1728 0.882544183 0.546943553 -1.154909670
552 SOLV HW2 1729 0.800154359 0.544027091 -1.013947919
553 SOLV OW 1730 -0.853649434 -1.188751973 -1.182509101
553 SOLV HW1 1731 -0.837561600 -1.153125340 -1.274552866
553 SOLV HW2 1732 -0.768729285 -1.227277821 -1.146392379
554 SOLV OW 1733 1.274815045 0.155211231 -0.205184855
554 SOLV HW1 1734 1.233225031 0.226126032 -0.148234125
554 SOLV HW2 1735 1.368455463 0.181685691 -0.228262811
555 SOLV OW 1736 1.203215312 -0.974312870 -1.408421835
555 SOLV HW1 1737 1.269867417 -0.981936370 -1.482590792
555 SOLV HW2 1738 1.182168969 -1.065456512 -1.373045496
556 SOLV OW 1739 1.237965621 1.346246151 -1.347892651
556 SOLV HW1 1740 1.169296145 1.416164407 -1.367828372
556 SOLV HW2 1741 1.194758444 1.269804023 -1.300026358
557 SOLV OW 1742 -0.037607157 0.267270436 1.187507242
557 SOLV HW1 1743 -0.089677501 0.324007150 1.251315441
557 SOLV HW2 1744 0.055600991 0.255524496 1.221801213
558 SOLV OW 1745 -1.189229873 -0.517369286 0.530195355
558 SOLV HW1 1746 -1.173878118 -0.529142269 0.628316218
558 SOLV HW2 1747 -1.163351026 -0.601170967 0.482138754
559 SOLV OW 1748 0.141765893 1.413439015 0.447679948
559 SOLV HW1 1749 0.151268657 1.510052217 0.471707530
559 SOLV HW2 1750 0.222077188 1.383932569 0.395899943
560 SOLV OW 1751 1.201578891 -0.091952221 -1.386247025
560 SOLV HW1 1752 1.127813920 -0.024773478 -1.379355192
560 SOLV HW2 1753 1.174574876 -0.176016117 -1.339281745
561 SOLV OW 1754 -0.453996811 -1.100106855 -0.748241675
561 SOLV HW1 1755 -0.354531842 -1.103864199 -0.738613978
561 SOLV HW2 1756 -0.492415903 -1.191151046 -0.732848121
562 SOLV OW 1757 1.125811364 0.989300095 0.580631744
562 SOLV HW1 1758 1.069859646 1.011122159 0.660590057
562 SOLV HW2 1759 1.206877399 1.047845511 0.579724871
563 SOLV OW 1760 -0.027462321 1.477059219 0.246018703
563 SOLV HW1 1761 0.027602798 1.439709185 0.320680379
563 SOLV HW2 1762 -0.062010652 1.402194655 0.189421499
564 SOLV OW 1763 -0.122002755 1.305279369 1.420965022
564 SOLV HW1 1764 -0.110546246 1.290409165 1.322742237
564 SOLV HW2 1765 -0.174099549 1.229498541 1.460274654
565 SOLV OW 1766 -1.217762380 0.601493299 1.543629493
565 SOLV HW1 1767 -1.266260884 0.531026829 1.491836591
565 SOLV HW2 1768 -1.261595913 0.690084057 1.528450921
566 SOLV OW 1769 -1.345533028 -1.408600324 -0.079618330
566 SOLV HW1 1770 -1.297766199 -1.321252439 -0.089039965
566 SOLV HW2 1771 -1.327538187 -1.446797434 0.011041723
567 SOLV OW 1772 -1.120855949 -0.125822550 -1.034913085
567 SOLV HW1 1773 -1.208278788 -0.171793505 -1.019232456
567 SOLV HW2 1774 -1.114984275 -0.044679800 -0.976746646
568 SOLV OW 1775 0.217003630 0.101227077 -0.893482050
568 SOLV HW1 1776 0.235142384 0.032063070 -0.823562196
568 SOLV HW2 1777 0.236449244 0.063118908 -0.983876358
569 SOLV OW 1778 0.187670114 1.300797575 -0.299396913
569 SOLV HW1 1779 0.189782404 1.370079386 -0.371489151
569 SOLV HW2 1780 0.153857251 1.214544530 -0.337063929
570 SOLV OW 1781 -0.671251251 0.424708835 -0.832459684
570 SOLV HW1 1782 -0.669723200 0.325368071 -0.821167092
570 SOLV HW2 1783 -0.683091432 0.447101363 -0.929190700
571 SOLV OW 1784 0.951884666 -0.882845576 0.115940256
571 SOLV HW1 1785 0.961287050 -0.804564339 0.177468518
571 SOLV HW2 1786 0.860248835 -0.921767382 0.125324162
572 SOLV OW 1787 -0.477163893 0.623073178 -0.867478573
572 SOLV HW1 1788 -0.476308191 0.583263469 -0.775738238
572 SOLV HW2 1789 -0.565839395 0.605905459 -0.910416772
573 SOLV OW 1790 -0.151449072 -1.109537068 -0.749946133
573 SOLV HW1 1791 -0.152419829 -1.186577293 -0.813707340
573 SOLV HW2 1792 -0.133549776 -1.143072444 -0.657444571
574 SOLV OW 1793 0.362112023 0.843391881 -1.497906406
574 SOLV HW1 1794 0.372961472 0.749066846 -1.529320087
574 SOLV HW2 1795 0.400889587 0.852627190 -1.406185691
575 SOLV OW 1796 -0.184018607 -0.029981781 0.222705207
575 SOLV HW1 1797 -0.094395198 -0.052220483 0.261087166
575 SOLV HW2 1798 -0.255455700 -0.055837334 0.287746043
576 SOLV OW 1799 -0.515032532 -0.056894842 0.588729230
576 SOLV HW1 1800 -0.476467549 -0.071594966 0.679815893
576 SOLV HW2 1801 -0.448140265 -0.083915961 0.519480592
577 SOLV OW 1802 0.491504064 1.363235542 -1.421190274
577 SOLV HW1 1803 0.434839275 1.404312514 -1.492630079
577 SOLV HW2 1804 0.445471507 1.282366834 -1.384542101
578 SOLV OW 1805 0.624511165 -1.298999453 -0.788902120
578 SOLV HW1 1806 0.540098040 -1.310874785 -0.736605307
578 SOLV HW2 1807 0.695889391 -1.357737442 -0.750738605
579 SOLV OW 1808 0.221076786 1.497573524 -0.771656237
579 SOLV HW1 1809 0.149481870 1.519605413 -0.837893575
579 SOLV HW2 1810 0.280487420 1.426436919 -0.809188500
580 SOLV OW 1811 1.459186494 1.081328092 -0.846178439
580 SOLV HW1 1812 1.449653787 1.064122883 -0.944233689
580 SOLV HW2 1813 1.472664806 0.994572030 -0.798288577
581 SOLV OW 1814 -0.330318018 0.588626075 -1.516862107
581 SOLV HW1 1815 -0.283529824 0.511668712 -1.473386697
581 SOLV HW2 1816 -0.268282919 0.633151675 -1.581443281
582 SOLV OW 1817 -1.102455353 0.581977114 -0.554819617
582 SOLV HW1 1818 -1.011088580 0.542349868 -0.545689728
582 SOLV HW2 1819 -1.096812023 0.667011712 -0.607152885
583 SOLV OW 1820 0.475285971 0.143132379 0.364243925
583 SOLV HW1 1821 0.426288912 0.228359729 0.382608229
583 SOLV HW2 1822 0.449347530 0.074487304 0.432190292
584 SOLV OW 1823 0.951978283 -1.407503729 -0.475025908
584 SOLV HW1 1824 0.871291718 -1.463883662 -0.457344662
584 SOLV HW2 1825 0.962318524 -1.340228260 -0.401754500
585 SOLV OW 1826 0.131883952 -0.553647260 -1.516840977
585 SOLV HW1 1827 0.122765161 -0.616564263 -1.594041540
585 SOLV HW2 1828 0.118238032 -0.603656420 -1.431315708
586 SOLV OW 1829 -1.377286927 -0.934005717 0.787769667
586 SOLV HW1 1830 -1.414298919 -1.014710476 0.741741026
586 SOLV HW2 1831 -1.284398009 -0.953351012 0.819378478
587 SOLV OW 1832 0.346816903 0.126402813 0.991787496
587 SOLV HW1 1833 0.376930627 0.204556219 1.046425755
587 SOLV HW2 1834 0.256790355 0.145742434 0.952783781
588 SOLV OW 1835 -0.290768567 1.163259344 0.887484404
588 SOLV HW1 1836 -0.263532021 1.250574582 0.847038254
588 SOLV HW2 1837 -0.238772658 1.147802084 0.971503477
589 SOLV OW 1838 -0.953547955 -0.986538537 -0.834165303
589 SOLV HW1 1839 -1.027205959 -0.989776881 -0.901735739
589 SOLV HW2 1840 -0.932323387 -1.079349105 -0.803547563
590 SOLV OW 1841 0.360121922 0.498541343 1.508837246
590 SOLV HW1 1842 0.330440457 0.420071884 1.563274799
590 SOLV HW2 1843 0.457957278 0.490680567 1.489649543
591 SOLV OW 1844 0.737544051 0.977272939 -0.281020979
591 SOLV HW1 1845 0.730569681 0.995460897 -0.379114315
591 SOLV HW2 1846 0.833734756 0.978438704 -0.253679009
592 SOLV OW 1847 0.802116840 0.018202504 0.091619468
592 SOLV HW1 1848 0.781620168 -0.047440358 0.164233243
592 SOLV HW2 1849 0.811090346 -0.030166443 0.004546285
593 SOLV OW 1850 -1.355855174 -0.112648903 -1.482693278
593 SOLV HW1 1851 -1.327137897 -0.038436577 -1.543272633
593 SOLV HW2 1852 -1.353095477 -0.199302825 -1.532546117
594 SOLV OW 1853 0.742507276 0.139330536 -0.578365357
594 SOLV HW1 1854 0.807590132 0.124793084 -0.652895077
594 SOLV HW2 1855 0.652078102 0.158735602 -0.616416074
595 SOLV OW 1856 -0.321545049 0.353449792 0.163468154
595 SOLV HW1 1857 -0.372593479 0.286649137 0.217630836
595 SOLV HW2 1858 -0.380398689 0.431661539 0.142949608
596 SOLV OW 1859 -0.453703124 0.322581263 -0.097733485
596 SOLV HW1 1860 -0.478350385 0.248885910 -0.160661178
596 SOLV HW2 1861 -0.516535875 0.322430025 -0.019948935
597 SOLV OW 1862 -0.847406112 0.977485930 -0.037399325
597 SOLV HW1 1863 -0.927461672 0.918654390 -0.025911083
597 SOLV HW2 1864 -0.866454383 1.067629136 0.001501315
598 SOLV OW 1865 -0.809611041 -1.173205847 1.460495082
598 SOLV HW1 1866 -0.842332922 -1.081402222 1.482928759
598 SOLV HW2 1867 -0.834558182 -1.195348221 1.366212580
599 SOLV OW 1868 -0.262232905 0.795978586 0.056424249
599 SOLV HW1 1869 -0.315400653 0.880670201 0.054949217
599 SOLV HW2 1870 -0.323010177 0.718811183 0.075211434
600 SOLV OW 1871 0.937806394 0.670830228 1.277581578
600 SOLV HW1 1872 0.981172263 0.714293473 1.356526326
600 SOLV HW2 1873 0.897426052 0.741037476 1.218910431
601 SOLV OW 1874 -1.369534036 -0.580721538 -0.672960742
601 SOLV HW1 1875 -1.312794733 -0.500207761 -0.690232553
601 SOLV HW2 1876 -1.431683872 -0.594822680 -0.750023352
602 SOLV OW 1877 1.059694885 -0.764102750 -0.527654512
602 SOLV HW1 1878 1.110047673 -0.839298121 -0.485089608
602 SOLV HW2 1879 0.991578892 -0.801154817 -0.590811129
603 SOLV OW 1880 1.178695023 -0.970057008 -0.430383683
603 SOLV HW1 1881 1.115031209 -1.036655916 -0.391481488
603 SOLV HW2 1882 1.226359624 -0.922428596 -0.356482810
604 SOLV OW 1883 -1.515592633 -0.243674707 1.211212212
604 SOLV HW1 1884 -1.537900377 -0.173367154 1.143675063
604 SOLV HW2 1885 -1.426695148 -0.283908648 1.189290623
605 SOLV OW 1886 0.612298106 0.246518304 -0.294821401
605 SOLV HW1 1887 0.705701504 0.211935931 -0.303866248
605 SOLV HW2 1888 0.573168032 0.214588943 -0.208500404
606 SOLV OW 1889 -0.151202652 -1.146255990 -1.409753537
606 SOLV HW1 1890 -0.113256185 -1.187942645 -1.327147289
606 SOLV HW2 1891 -0.125312109 -1.200775927 -1.489494957
607 SOLV OW 1892 0.237160838 -1.062740106 -0.314080249
607 SOLV HW1 1893 0.216349495 -1.158317609 -0.334907798
607 SOLV HW2 1894 0.152143854 -1.013369085 -0.295736824
608 SOLV OW 1895 0.904840280 0.082205779 -0.780638091
608 SOLV HW1 1896 0.853468607 0.023138297 -0.842864083
608 SOLV HW2 1897 1.002280583 0.076924717 -0.802537277
609 SOLV OW 1898 -0.775827722 0.654957564 -0.375075935
609 SOLV HW1 1899 -0.809067361 0.591211805 -0.305551370
609 SOLV HW2 1900 -0.785573422 0.748722020 -0.341681289
610 SOLV OW 1901 1.256462200 0.429103345 0.384439666
610 SOLV HW1 1902 1.343906988 0.440799116 0.431537839
610 SOLV HW2 1903 1.213571200 0.343903655 0.414489208
611 SOLV OW 1904 0.849761637 1.444597599 -0.577511392
611 SOLV HW1 1905 0.941881377 1.435333062 -0.615324569
611 SOLV HW2 1906 0.837384168 1.378610972 -0.503388240
612 SOLV OW 1907 0.031190140 -1.332897103 1.302477145
612 SOLV HW1 1908 0.029624371 -1.304019877 1.398204593
612 SOLV HW2 1909 -0.060864789 -1.325812902 1.264036526
613 SOLV OW 1910 -1.068435171 -0.930556650 1.216113185
613 SOLV HW1 1911 -1.010964452 -1.012324125 1.219478853
613 SOLV HW2 1912 -1.162710668 -0.956925553 1.195695012
614 SOLV OW 1913 -1.454618747 0.258558934 -1.441305593
614 SOLV HW1 1914 -1.468006811 0.333206699 -1.376110212
614 SOLV HW2 1915 -1.492040013 0.173981584 -1.403250071
615 SOLV OW 1916 1.042032837 1.024262180 -1.429596010
615 SOLV HW1 1917 0.964488537 1.073169209 -1.389657128
615 SOLV HW2 1918 1.122838752 1.037381190 -1.372164435
616 SOLV OW 1919 -0.392682463 -0.634737151 0.700233197
616 SOLV HW1 1920 -0.432049092 -0.590895024 0.619426205
616 SOLV HW2 1921 -0.293629345 -0.642625449 0.688929734
617 SOLV OW 1922 0.280905935 -0.759390886 -0.247044768
617 SOLV HW1 1923 0.290570745 -0.857930559 -0.261128641
617 SOLV HW2 1924 0.183845190 -0.735347416 -0.245322544
618 SOLV OW 1925 0.833849349 0.100260093 -0.273201536
618 SOLV HW1 1926 0.809779797 0.004875725 -0.255194887
618 SOLV HW2 1927 0.914696745 0.103693787 -0.331968766
619 SOLV OW 1928 -1.213105462 -1.189693745 -0.076971045
619 SOLV HW1 1929 -1.218534784 -1.138463071 0.008746234
619 SOLV HW2 1930 -1.210618364 -1.125803534 -0.153869479
620 SOLV OW 1931 1.447187903 1.304461552 -0.434902761
620 SOLV HW1 1932 1.440131919 1.288624021 -0.533396346
620 SOLV HW2 1933 1.401926295 1.390563446 -0.411677458
621 SOLV OW 1934 -1.034815961 -1.520707781 0.910626048
621 SOLV HW1 1935 -1.052369416 -1.423521889 0.894852251
621 SOLV HW2 1936 -0.942808277 -1.543539763 0.878762792
622 SOLV OW 1937 -0.798291197 0.849117234 1.173350854
622 SOLV HW1 1938 -0.825593286 0.855050451 1.077324070
622 SOLV HW2 1939 -0.879798278 0.850254276 1.231290363
623 SOLV OW 1940 0.475528250 0.648404246 0.560283599
623 SOLV HW1 1941 0.401862432 0.696116771 0.512342775
623 SOLV HW2 1942 0.511073356 0.706795643 0.633279699
624 SOLV OW 1943 -1.320821020 -1.508305572 0.188884749
624 SOLV HW1 1944 -1.223073003 -1.487696265 0.184165909
624 SOLV HW2 1945 -1.334767300 -1.591534118 0.242551593
625 SOLV OW 1946 0.694788952 1.276306092 -0.954137094
625 SOLV HW1 1947 0.793844995 1.276381024 -0.967899312
625 SOLV HW2 1948 0.670761535 1.344050426 -0.884603948
626 SOLV OW 1949 -0.994693596 -0.982498088 0.073044204
626 SOLV HW1 1950 -0.921358008 -0.958464303 0.136625016
626 SOLV HW2 1951 -1.076473229 -1.008073316 0.124582681
627 SOLV OW 1952 -1.296552140 -1.448465769 -0.650378186
627 SOLV HW1 1953 -1.317213050 -1.355667239 -0.619341547
627 SOLV HW2 1954 -1.380501115 -1.492320153 -0.682487456
628 SOLV OW 1955 0.735299600 -1.226509620 -0.251258031
628 SOLV HW1 1956 0.729017320 -1.320537202 -0.284737512
628 SOLV HW2 1957 0.715938944 -1.224709850 -0.153159100
629 SOLV OW 1958 0.390640467 -0.373869147 -1.244719646
629 SOLV HW1 1959 0.427442189 -0.298360306 -1.190444385
629 SOLV HW2 1960 0.317725677 -0.419272547 -1.193497384
630 SOLV OW 1961 -1.348398797 0.522395979 -0.705711967
630 SOLV HW1 1962 -1.383172455 0.487417682 -0.618720909
630 SOLV HW2 1963 -1.353433121 0.622269527 -0.705853884
631 SOLV OW 1964 0.080319326 0.960824873 0.879946994
631 SOLV HW1 1965 0.070399260 0.986295800 0.783746402
631 SOLV HW2 1966 0.007104694 1.003145081 0.933334848
632 SOLV OW 1967 1.384458529 -1.262460917 0.794081116
632 SOLV HW1 1968 1.305907132 -1.300927411 0.745585439
632 SOLV HW2 1969 1.363955252 -1.168478916 0.821444623
633 SOLV OW 1970 -0.554646096 0.480395800 0.128791998
633 SOLV HW1 1971 -0.552983422 0.511686253 0.033817653
633 SOLV HW2 1972 -0.641302592 0.506918333 0.171090357
634 SOLV OW 1973 -0.365436235 -1.291575545 0.700339327
634 SOLV HW1 1974 -0.316016768 -1.204662646 0.702337721
634 SOLV HW2 1975 -0.446058351 -1.282710121 0.641845149
635 SOLV OW 1976 0.209616530 -0.155979287 1.001834833
635 SOLV HW1 1977 0.263752555 -0.237514601 0.981342849
635 SOLV HW2 1978 0.254697810 -0.075466069 0.963312640
636 SOLV OW 1979 -0.297554556 -0.808751356 -1.538681878
636 SOLV HW1 1980 -0.266803259 -0.815497257 -1.443766222
636 SOLV HW2 1981 -0.219807995 -0.822886543 -1.599966126
637 SOLV OW 1982 -0.619502529 0.543885631 1.319964155
637 SOLV HW1 1983 -0.677964158 0.515912476 1.243794902
637 SOLV HW2 1984 -0.542055946 0.596567187 1.284911560
638 SOLV OW 1985 -0.679129957 -1.447000707 0.014831156
638 SOLV HW1 1986 -0.707614972 -1.387905145 0.090315947
638 SOLV HW2 1987 -0.753122719 -1.510446392 -0.007558930
639 SOLV OW 1988 1.014986824 0.262011490 -0.502093901
639 SOLV HW1 1989 0.918320303 0.247962463 -0.523539657
639 SOLV HW2 1990 1.025645233 0.346984414 -0.450445085
640 SOLV OW 1991 -1.374634300 0.185162371 -0.596215867
640 SOLV HW1 1992 -1.406594135 0.159796685 -0.687521279
640 SOLV HW2 1993 -1.452864990 0.210961334 -0.539507799
641 SOLV OW 1994 1.305761714 -0.274879121 1.248945043
641 SOLV HW1 1995 1.390308883 -0.250361294 1.296387452
641 SOLV HW2 1996 1.265269508 -0.192866787 1.208516731
642 SOLV OW 1997 1.503260826 0.455420944 0.501990233
642 SOLV HW1 1998 1.513286456 0.516579027 0.580482544
642 SOLV HW2 1999 1.559125014 0.489088123 0.426178038
643 SOLV OW 2000 -0.866425692 0.279592156 0.083850495
643 SOLV HW1 2001 -0.834609864 0.359232018 0.135283716
643 SOLV HW2 2002 -0.791988561 0.213318449 0.075665175
644 SOLV OW 2003 0.517130781 -0.540329708 1.269627266
644 SOLV HW1 2004 0.455896386 -0.590642569 1.208630017
644 SOLV HW2 2005 0.502239552 -0.570257818 1.363883284
645 SOLV OW 2006 -0.634654959 -0.348501515 -0.700099532
645 SOLV HW1 2007 -0.656431145 -0.251027670 -0.694963731
645 SOLV HW2 2008 -0.535370854 -0.360419586 -0.701643452
646 SOLV OW 2009 0.983798974 0.561646660 0.609705076
646 SOLV HW1 2010 1.063722535 0.514596247 0.647128251
646 SOLV HW2 2011 0.966546149 0.529172346 0.516701676
647 SOLV OW 2012 -1.203504159 -1.362970410 1.275354782
647 SOLV HW1 2013 -1.165210156 -1.319064465 1.356631862
647 SOLV HW2 2014 -1.239660949 -1.452892681 1.299987317
648 SOLV OW 2015 -0.210766041 1.244237091 -0.057121518
648 SOLV HW1 2016 -0.309791294 1.255493710 -0.048826260
648 SOLV HW2 2017 -0.190823366 1.158056895 -0.103776267
649 SOLV OW 2018 -1.113523042 0.181673321 -0.008495740
649 SOLV HW1 2019 -1.021168388 0.212560490 0.014267163
649 SOLV HW2 2020 -1.128263510 0.190483684 -0.107017846
650 SOLV OW 2021 1.413788780 -1.087944062 -0.868486715
650 SOLV HW1 2022 1.490924722 -1.124621420 -0.816459212
650 SOLV HW2 2023 1.384175477 -1.155374209 -0.936149097
651 SOLV OW 2024 0.548673504 -0.798173674 -1.421015829
651 SOLV HW1 2025 0.552532527 -0.731506570 -1.495465068
651 SOLV HW2 2026 0.552543940 -0.890656300 -1.458884098
652 SOLV OW 2027 0.365439588 -0.409282511 1.055781931
652 SOLV HW1 2028 0.288460417 -0.401312358 1.119123872
652 SOLV HW2 2029 0.448524331 -0.375313403 1.099878057
653 SOLV OW 2030 0.556095408 1.101014424 -0.662200539
653 SOLV HW1 2031 0.571597771 1.195697638 -0.633975069
653 SOLV HW2 2032 0.628307230 1.042839477 -0.624743395
654 SOLV OW 2033 0.257600566 -1.425460810 1.228207448
654 SOLV HW1 2034 0.317327975 -1.346500049 1.214076360
654 SOLV HW2 2035 0.163504472 -1.394158780 1.241164717
655 SOLV OW 2036 1.009372017 -0.532553882 1.221402626
655 SOLV HW1 2037 0.962629414 -0.446801704 1.199869256
655 SOLV HW2 2038 0.958916433 -0.608992442 1.181236426
656 SOLV OW 2039 1.437330206 0.632101495 0.697402609
656 SOLV HW1 2040 1.379612823 0.684415782 0.634682246
656 SOLV HW2 2041 1.512879134 0.689555518 0.728918531
657 SOLV OW 2042 1.200956144 0.611604574 -1.111523589
657 SOLV HW1 2043 1.270814590 0.682996784 -1.116480652
657 SOLV HW2 2044 1.161174782 0.597566006 -1.202198203
658 SOLV OW 2045 1.195507009 -0.084992161 0.063463016
658 SOLV HW1 2046 1.237907268 -0.108636888 -0.023972981
658 SOLV HW2 2047 1.135942647 -0.159742761 0.092895363
659 SOLV OW 2048 0.823084945 -0.609328595 -0.154929317
659 SOLV HW1 2049 0.893165488 -0.673075661 -0.186947405
659 SOLV HW2 2050 0.822260599 -0.608208004 -0.054938543
660 SOLV OW 2051 -1.315059752 1.338922532 -0.488219074
660 SOLV HW1 2052 -1.216945325 1.344946607 -0.506632509
660 SOLV HW2 2053 -1.329507869 1.313591052 -0.392556627
661 SOLV OW 2054 0.455097591 0.494111003 -0.063762571
661 SOLV HW1 2055 0.551508154 0.489550460 -0.037603539
661 SOLV HW2 2056 0.447920521 0.502752052 -0.163130360
662 SOLV OW 2057 0.625273463 -0.776190349 -1.035554542
662 SOLV HW1 2058 0.605615612 -0.708218708 -1.106230132
662 SOLV HW2 2059 0.619628484 -0.867898901 -1.075042329
663 SOLV OW 2060 1.285996071 0.145922686 0.288952641
663 SOLV HW1 2061 1.362508522 0.175475913 0.231733385
663 SOLV HW2 2062 1.214221505 0.107084244 0.231146774
664 SOLV OW 2063 0.705389581 0.483875585 0.063693984
664 SOLV HW1 2064 0.693760272 0.562315006 0.124634835
664 SOLV HW2 2065 0.703885482 0.399492381 0.117349444
665 SOLV OW 2066 0.506903405 1.009378206 0.455773726
665 SOLV HW1 2067 0.542144333 0.959018227 0.534665219
665 SOLV HW2 2068 0.406975499 1.011222159 0.459359577
666 SOLV OW 2069 0.255745388 -0.849504545 -0.675457250
666 SOLV HW1 2070 0.257899997 -0.865767278 -0.774111835
666 SOLV HW2 2071 0.272837323 -0.935391058 -0.627155346
667 SOLV OW 2072 0.530067893 0.095177122 -0.078629179
667 SOLV HW1 2073 0.461518881 0.113638532 -0.008199488
667 SOLV HW2 2074 0.617177365 0.072339413 -0.035149653
668 SOLV OW 2075 -1.221498816 -0.459212967 -1.497954853
668 SOLV HW1 2076 -1.179701214 -0.410255489 -1.421416799
668 SOLV HW2 2077 -1.221319280 -0.400661656 -1.579032468
669 SOLV OW 2078 -0.555193186 -1.357222335 -0.766967497
669 SOLV HW1 2079 -0.641762445 -1.331437913 -0.724037372
669 SOLV HW2 2080 -0.571918789 -1.427606862 -0.836020410
670 SOLV OW 2081 -0.676959430 0.452739009 0.778819050
670 SOLV HW1 2082 -0.655452893 0.515656440 0.853524293
670 SOLV HW2 2083 -0.633044497 0.364521341 0.795880128
671 SOLV OW 2084 0.128316509 -1.150380274 -0.836225774
671 SOLV HW1 2085 0.191423928 -1.148839474 -0.913792139
671 SOLV HW2 2086 0.066068103 -1.072348681 -0.842364696
672 SOLV OW 2087 -0.832027119 1.276184188 0.952854376
672 SOLV HW1 2088 -0.906350505 1.337443113 0.925957008
672 SOLV HW2 2089 -0.747810993 1.303580975 0.906388779
673 SOLV OW 2090 0.092608975 -1.403362577 0.893345618
673 SOLV HW1 2091 0.002312381 -1.386340566 0.932819909
673 SOLV HW2 2092 0.159426748 -1.341889921 0.935273437
674 SOLV OW 2093 0.564823735 -0.401216640 -0.013460623
674 SOLV HW1 2094 0.555285729 -0.488212756 -0.061858381
674 SOLV HW2 2095 0.545497605 -0.325846663 -0.076289191
675 SOLV OW 2096 1.049478261 -0.126713958 -1.150721243
675 SOLV HW1 2097 0.973481224 -0.067309562 -1.177131895
675 SOLV HW2 2098 1.127075174 -0.070653212 -1.121773765
676 SOLV OW 2099 0.350517271 -0.626264559 -0.931907811
676 SOLV HW1 2100 0.317111017 -0.720528112 -0.932381580
676 SOLV HW2 2101 0.449899154 -0.626085170 -0.943088827
677 SOLV OW 2102 -0.405294001 -0.612508805 -1.160579602
677 SOLV HW1 2103 -0.451827765 -0.568340435 -1.237296702
677 SOLV HW2 2104 -0.325992673 -0.663178799 -1.194424587
678 SOLV OW 2105 1.017483034 -0.287064465 0.129936525
678 SOLV HW1 2106 1.016943109 -0.353081893 0.205055793
678 SOLV HW2 2107 0.958652997 -0.320106274 0.056120656
679 SOLV OW 2108 1.481277417 -1.446739857 0.316569184
679 SOLV HW1 2109 1.425283557 -1.366221973 0.296994194
679 SOLV HW2 2110 1.524470664 -1.478597650 0.232183479
680 SOLV OW 2111 -1.286152909 1.046217398 0.135045977
680 SOLV HW1 2112 -1.370394205 1.026267382 0.084972992
680 SOLV HW2 2113 -1.260828385 1.141918749 0.120846329
681 SOLV OW 2114 -1.328761988 -1.099571231 -0.523964781
681 SOLV HW1 2115 -1.305711592 -1.118227081 -0.619475836
681 SOLV HW2 2116 -1.358854075 -1.004629970 -0.514889141
682 SOLV OW 2117 0.723597751 -1.502523998 -0.361509460
682 SOLV HW1 2118 0.737412025 -1.564460831 -0.284223442
682 SOLV HW2 2119 0.630891767 -1.513974951 -0.397210528
683 SOLV OW 2120 0.522327615 0.487526832 -1.127521916
683 SOLV HW1 2121 0.490289046 0.482559625 -1.222128809
683 SOLV HW2 2122 0.589560753 0.415294856 -1.111280341
684 SOLV OW 2123 0.598406938 1.429356256 -1.196484752
684 SOLV HW1 2124 0.543331112 1.407048806 -1.276927633
684 SOLV HW2 2125 0.620518866 1.345355622 -1.146916842
685 SOLV OW 2126 1.035925597 -1.440803654 -1.307362771
685 SOLV HW1 2127 1.090233391 -1.467146689 -1.387102912
685 SOLV HW2 2128 1.042718281 -1.511983035 -1.237440558
686 SOLV OW 2129 1.218058496 -0.896854913 0.644481700
686 SOLV HW1 2130 1.124181785 -0.931332103 0.644588138
686 SOLV HW2 2131 1.272083330 -0.947108483 0.711990233
687 SOLV OW 2132 0.924975878 -0.207036570 1.515280945
687 SOLV HW1 2133 0.981532543 -0.270808407 1.462988802
687 SOLV HW2 2134 0.836174139 -0.196774001 1.470458831
688 SOLV OW 2135 -1.465851919 1.039529384 -1.259185352
688 SOLV HW1 2136 -1.515347842 0.974244284 -1.316527661
688 SOLV HW2 2137 -1.428917511 1.112776758 -1.316375926
689 SOLV OW 2138 0.740621795 -0.119249157 0.894203316
689 SOLV HW1 2139 0.821493698 -0.062701398 0.910441698
689 SOLV HW2 2140 0.714709036 -0.165466902 0.979020783
690 SOLV OW 2141 -0.906602811 0.857667137 0.685819981
690 SOLV HW1 2142 -0.940019691 0.918549295 0.613858648
690 SOLV HW2 2143 -0.808514935 0.842411720 0.673661430
691 SOLV OW 2144 -0.954918814 -0.295232450 0.045479647
691 SOLV HW1 2145 -1.030470462 -0.360523392 0.039953122
691 SOLV HW2 2146 -0.964126032 -0.227376725 -0.027405511
692 SOLV OW 2147 -0.704651228 -0.338376064 -0.279265196
692 SOLV HW1 2148 -0.617389379 -0.304961111 -0.314913826
692 SOLV HW2 2149 -0.764106334 -0.364097329 -0.355458205
693 SOLV OW 2150 1.368777599 0.814106955 0.142480520
693 SOLV HW1 2151 1.432749649 0.775964087 0.209220579
693 SOLV HW2 2152 1.275420441 0.806007719 0.177413282
694 SOLV OW 2153 -0.303845039 0.085587829 1.362810498
694 SOLV HW1 2154 -0.325081178 0.108363634 1.457848026
694 SOLV HW2 2155 -0.339767676 0.156928999 1.302630386
695 SOLV OW 2156 -0.946442193 1.095822908 -0.671180445
695 SOLV HW1 2157 -0.860522698 1.146641590 -0.677252550
695 SOLV HW2 2158 -0.976223355 1.069640569 -0.762990550
696 SOLV OW 2159 0.911966187 1.454116889 0.248902060
696 SOLV HW1 2160 0.900885366 1.369608575 0.196580195
696 SOLV HW2 2161 0.993671819 1.502376081 0.217323087
697 SOLV OW 2162 0.922169251 -0.649327899 0.492883781
697 SOLV HW1 2163 0.840108597 -0.612767173 0.536809740
697 SOLV HW2 2164 0.945434212 -0.737505968 0.533913768
698 SOLV OW 2165 -0.865712808 -0.479269428 -1.238781254
698 SOLV HW1 2166 -0.830283462 -0.521747730 -1.322102205
698 SOLV HW2 2167 -0.789370179 -0.452512548 -1.179977622
699 SOLV OW 2168 0.886660710 -1.303271450 1.059996036
699 SOLV HW1 2169 0.899619360 -1.402383216 1.063228284
699 SOLV HW2 2170 0.803831893 -1.282177052 1.008073562
700 SOLV OW 2171 0.664744857 0.272695999 0.921765739
700 SOLV HW1 2172 0.570140794 0.244676300 0.905431530
700 SOLV HW2 2173 0.683495754 0.357346340 0.871921546
701 SOLV OW 2174 -0.260689826 -1.437435704 0.471788056
701 SOLV HW1 2175 -0.352200983 -1.476849286 0.463169343
701 SOLV HW2 2176 -0.249699451 -1.398421219 0.563215723
702 SOLV OW 2177 1.016690217 0.033536585 1.373323292
702 SOLV HW1 2178 0.948847939 0.106983751 1.371135752
702 SOLV HW2 2179 0.977698821 -0.046481476 1.418914981
703 SOLV OW 2180 -0.945634024 -0.948114395 1.520889539
703 SOLV HW1 2181 -1.027838705 -0.953936354 1.577549337
703 SOLV HW2 2182 -0.962049206 -0.885427604 1.444714488
704 SOLV OW 2183 -0.657890606 -0.369620155 1.299013896
704 SOLV HW1 2184 -0.688603111 -0.312685845 1.375272118
704 SOLV HW2 2185 -0.735493094 -0.390040465 1.239323943
705 SOLV OW 2186 -1.163940966 0.524016729 0.056977119
705 SOLV HW1 2187 -1.193408327 0.430611592 0.036757440
705 SOLV HW2 2188 -1.112539344 0.560298258 -0.020761080
706 SOLV OW 2189 -0.438302638 0.165752255 0.301704218
706 SOLV HW1 2190 -0.494544432 0.129362194 0.375962348
706 SOLV HW2 2191 -0.359968992 0.105439395 0.286609371
707 SOLV OW 2192 1.004818217 0.919596586 -0.178361056
707 SOLV HW1 2193 1.037908343 1.013742369 -0.171795470
707 SOLV HW2 2194 1.066626013 0.866526749 -0.236368377
708 SOLV OW 2195 -0.913273044 -0.674238267 1.310084986
708 SOLV HW1 2196 -0.888484297 -0.667682380 1.406751763
708 SOLV HW2 2197 -0.836406162 -0.712048976 1.258474061
709 SOLV OW 2198 0.161437797 -0.852955089 1.320210090
709 SOLV HW1 2199 0.063829577 -0.863264545 1.301066405
709 SOLV HW2 2200 0.205256492 -0.942838048 1.319177712
710 SOLV OW 2201 0.276338526 -1.137700712 1.326165224
710 SOLV HW1 2202 0.197369932 -1.194284011 1.349913822
710 SOLV HW2 2203 0.338677636 -1.133056188 1.404228557
711 SOLV OW 2204 -0.611065508 0.142848412 -1.335920013
711 SOLV HW1 2205 -0.527834888 0.115380660 -1.384086073
711 SOLV HW2 2206 -0.686589258 0.149637159 -1.401125626
712 SOLV OW 2207 -1.172281743 0.347711239 0.743187592
712 SOLV HW1 2208 -1.214159490 0.261567901 0.714425733
712 SOLV HW2 2209 -1.072971572 0.341447022 0.733190103
713 SOLV OW 2210 -0.423173040 -1.409120194 1.388285024
713 SOLV HW1 2211 -0.389626705 -1.473369202 1.319376060
713 SOLV HW2 2212 -0.408784576 -1.315459463 1.356312413
714 SOLV OW 2213 -0.355622361 -0.651159718 1.151497371
714 SOLV HW1 2214 -0.412917686 -0.620545280 1.075461451
714 SOLV HW2 2215 -0.398883483 -0.625948952 1.238067248
715 SOLV OW 2216 -1.321317398 -1.361018096 -1.006610637
715 SOLV HW1 2217 -1.307659930 -1.432937519 -1.074737005
715 SOLV HW2 2218 -1.236378954 -1.309519347 -0.995059454
716 SOLV OW 2219 1.039223701 0.988716113 0.301178242
716 SOLV HW1 2220 1.080542446 0.972193164 0.390742832
716 SOLV HW2 2221 0.942466116 0.963574003 0.303982003
717 SOLV OW 2222 -0.825626696 -1.366506969 -0.276478837
717 SOLV HW1 2223 -0.838880335 -1.268477454 -0.261828055
717 SOLV HW2 2224 -0.741649651 -1.395954068 -0.230841099
718 SOLV OW 2225 -0.415475616 -1.128128736 -1.459901214
718 SOLV HW1 2226 -0.472005588 -1.058696059 -1.415345859
718 SOLV HW2 2227 -0.320649617 -1.118299120 -1.429686331
719 SOLV OW 2228 0.667891229 1.146697396 1.310493083
719 SOLV HW1 2229 0.602648620 1.180704129 1.378232264
719 SOLV HW2 2230 0.711313413 1.224042547 1.264300056
720 SOLV OW 2231 -0.965546310 1.277225998 0.619087758
720 SOLV HW1 2232 -0.979100063 1.250854599 0.523576227
720 SOLV HW2 2233 -1.032719942 1.230335298 0.676450916
721 SOLV OW 2234 -1.541835235 0.689862524 -0.305807220
721 SOLV HW1 2235 -1.444423658 0.672423876 -0.291364803
721 SOLV HW2 2236 -1.579514034 0.736659040 -0.225857264
722 SOLV OW 2237 -1.110920214 -1.489796088 0.488183591
722 SOLV HW1 2238 -1.131169583 -1.418312615 0.555117935
722 SOLV HW2 2239 -1.091461737 -1.447851372 0.399515115
723 SOLV OW 2240 0.671328669 -1.001829088 1.127752024
723 SOLV HW1 2241 0.598799761 -0.959591133 1.182117816
723 SOLV HW2 2242 0.743733903 -1.034525421 1.188485227
724 SOLV OW 2243 0.995646679 -0.033809908 0.796468217
724 SOLV HW1 2244 1.052011027 -0.086847218 0.733127294
724 SOLV HW2 2245 0.969446172 0.052573500 0.753417983
725 SOLV OW 2246 -0.506132281 0.522849039 -0.394396372
725 SOLV HW1 2247 -0.603617421 0.544824198 -0.390425139
725 SOLV HW2 2248 -0.470119256 0.514221130 -0.301495412
726 SOLV OW 2249 -1.133359655 -1.087446762 -0.326112053
726 SOLV HW1 2250 -1.199359382 -1.127014074 -0.389988898
726 SOLV HW2 2251 -1.043509602 -1.082644027 -0.369766417
727 SOLV OW 2252 0.077198366 -1.311114564 -0.354880936
727 SOLV HW1 2253 0.044371690 -1.294676091 -0.261852834
727 SOLV HW2 2254 0.101881051 -1.407506702 -0.364948570
728 SOLV OW 2255 -1.455019818 -0.526109659 1.109220089
728 SOLV HW1 2256 -1.440692628 -0.620016116 1.077972704
728 SOLV HW2 2257 -1.473314102 -0.526091334 1.207523151
729 SOLV OW 2258 -0.223433650 0.235288319 -1.267299748
729 SOLV HW1 2259 -0.165284471 0.192357405 -1.198179211
729 SOLV HW2 2260 -0.180548738 0.319880618 -1.299033741
730 SOLV OW 2261 1.415563175 -0.842987365 0.272490156
730 SOLV HW1 2262 1.504590772 -0.831383818 0.228434724
730 SOLV HW2 2263 1.393813583 -0.761164211 0.325719999
731 SOLV OW 2264 -1.513161885 1.039126588 0.573593894
731 SOLV HW1 2265 -1.540826274 0.970794471 0.506013185
731 SOLV HW2 2266 -1.446944372 0.998677297 0.636686595
732 SOLV OW 2267 1.498759001 -1.218383614 0.052292622
732 SOLV HW1 2268 1.440542744 -1.207899178 -0.028336599
732 SOLV HW2 2269 1.441998762 -1.218075951 0.134622876
733 SOLV OW 2270 -1.488534120 -0.072940140 0.509640171
733 SOLV HW1 2271 -1.428014555 -0.127570905 0.451721429
733 SOLV HW2 2272 -1.539749827 -0.008487965 0.452855446
734 SOLV OW 2273 0.211793136 -1.089879243 1.067994722
734 SOLV HW1 2274 0.115302235 -1.070667223 1.050091205
734 SOLV HW2 2275 0.222936280 -1.118630002 1.163122606
735 SOLV OW 2276 1.544664999 -0.066232822 0.687873002
735 SOLV HW1 2277 1.605744832 -0.036468021 0.761254726
735 SOLV HW2 2278 1.451208947 -0.072704467 0.722883071
736 SOLV OW 2279 -0.503452907 -1.542035413 0.331830724
736 SOLV HW1 2280 -0.599623500 -1.566165959 0.318766411
736 SOLV HW2 2281 -0.465664164 -1.507958458 0.245735001
737 SOLV OW 2282 1.095996076 0.497401373 -0.735617748
737 SOLV HW1 2283 1.167373451 0.558308783 -0.770221688
737 SOLV HW2 2284 1.093282747 0.502604434 -0.635780634
738 SOLV OW 2285 0.784397501 -0.581685459 0.145910787
738 SOLV HW1 2286 0.858081749 -0.523388463 0.180169459
738 SOLV HW2 2287 0.696270378 -0.537833792 0.163579810
739 SOLV OW 2288 0.441813041 -1.370715696 -0.996026717
739 SOLV HW1 2289 0.462342963 -1.439009055 -1.066144745
739 SOLV HW2 2290 0.525839712 -1.346502037 -0.947496447
740 SOLV OW 2291 -0.118740465 -1.298760668 -0.916477957
740 SOLV HW1 2292 -0.173336677 -1.307871627 -0.999771582
740 SOLV HW2 2293 -0.023551061 -1.280210787 -0.940899254
741 SOLV OW 2294 1.456490362 -0.262162582 0.407904441
741 SOLV HW1 2295 1.547787054 -0.236021181 0.439260914
741 SOLV HW2 2296 1.440485075 -0.224338472 0.316718663
742 SOLV OW 2297 -0.365902047 0.522798161 1.515070022
742 SOLV HW1 2298 -0.448403275 0.469125960 1.532798624
742 SOLV HW2 2299 -0.391365458 0.617430481 1.495125550
743 SOLV OW 2300 -1.480864587 -0.958230826 -1.166395920
743 SOLV HW1 2301 -1.527246052 -1.043322617 -1.141700313
743 SOLV HW2 2302 -1.460671704 -0.906029829 -1.083516547
744 SOLV OW 2303 -0.596066996 -0.122742221 1.044305081
744 SOLV HW1 2304 -0.555235132 -0.212454779 1.061218443
744 SOLV HW2 2305 -0.526048085 -0.060248289 1.009758705
745 SOLV OW 2306 0.354871273 -0.915812585 -0.917342458
745 SOLV HW1 2307 0.337742797 -0.998584371 -0.970795970
745 SOLV HW2 2308 0.444057623 -0.922881416 -0.872647998
746 SOLV OW 2309 -1.035865100 0.007967266 0.639182992
746 SOLV HW1 2310 -1.108735811 0.027101214 0.704951991
746 SOLV HW2 2311 -1.070864143 0.019934414 0.546266208
747 SOLV OW 2312 0.661768895 1.060254443 0.866374386
747 SOLV HW1 2313 0.733919522 1.121001403 0.833115204
747 SOLV HW2 2314 0.645047428 1.078343864 0.963303104
748 SOLV OW 2315 -1.104024105 -0.996289489 0.558356673
748 SOLV HW1 2316 -1.164668554 -1.075773725 0.560920911
748 SOLV HW2 2317 -1.101085966 -0.953701401 0.648798133
749 SOLV OW 2318 0.330223869 1.315702871 0.288622341
749 SOLV HW1 2319 0.343496783 1.401656059 0.239250723
749 SOLV HW2 2320 0.319508628 1.240940408 0.223068887
750 SOLV OW 2321 0.379643512 -0.276400505 0.616052867
750 SOLV HW1 2322 0.402869056 -0.204296482 0.681346813
750 SOLV HW2 2323 0.281466397 -0.273885952 0.597171379
751 SOLV OW 2324 -0.012935470 -1.298983099 -1.201103968
751 SOLV HW1 2325 0.068408613 -1.244147717 -1.220547919
751 SOLV HW2 2326 0.011750122 -1.395889188 -1.199783882
752 SOLV OW 2327 0.426356825 -0.851387209 0.048963361
752 SOLV HW1 2328 0.462488054 -0.813115761 0.134000715
752 SOLV HW2 2329 0.345161368 -0.906238150 0.068966315
753 SOLV OW 2330 -0.158601951 0.910004584 0.477187375
753 SOLV HW1 2331 -0.122684758 0.975615932 0.543570058
753 SOLV HW2 2332 -0.095792467 0.903283923 0.399654496
754 SOLV OW 2333 -0.473131462 -0.575758723 1.402740896
754 SOLV HW1 2334 -0.515231486 -0.653844726 1.448914365
754 SOLV HW2 2335 -0.539425880 -0.501157430 1.396302345
755 SOLV OW 2336 -1.394635886 1.328441592 -1.258631412
755 SOLV HW1 2337 -1.299926502 1.359807959 -1.265444033
755 SOLV HW2 2338 -1.449270424 1.371632958 -1.330392566
756 SOLV OW 2339 1.226038809 0.290297878 1.116708180
756 SOLV HW1 2340 1.156981252 0.279426844 1.188226045
756 SOLV HW2 2341 1.192183318 0.353573796 1.047052932
757 SOLV OW 2342 0.629911830 0.545393734 -0.662618992
757 SOLV HW1 2343 0.601209689 0.579671296 -0.752079335
757 SOLV HW2 2344 0.611621778 0.447224468 -0.657125504
758 SOLV OW 2345 -1.484543855 0.174799764 -0.820439475
758 SOLV HW1 2346 -1.456686989 0.177466409 -0.916444548
758 SOLV HW2 2347 -1.540723981 0.254928296 -0.799818224
759 SOLV OW 2348 0.827604773 0.599737878 -0.165146775
759 SOLV HW1 2349 0.790770550 0.568593038 -0.077538647
759 SOLV HW2 2350 0.883363234 0.681488445 -0.150668846
760 SOLV OW 2351 -0.412843880 0.277970548 -0.583787481
760 SOLV HW1 2352 -0.510235629 0.258963047 -0.596184688
760 SOLV HW2 2353 -0.395034443 0.374479324 -0.602996084
761 SOLV OW 2354 -1.270125911 0.370677110 -0.211934596
761 SOLV HW1 2355 -1.201127835 0.441309110 -0.196051352
761 SOLV HW2 2356 -1.262855822 0.337564563 -0.306021932
762 SOLV OW 2357 1.072116651 0.481652700 0.928118138
762 SOLV HW1 2358 1.117100922 0.449694961 0.844710226
762 SOLV HW2 2359 0.997753814 0.419153182 0.951902910
763 SOLV OW 2360 -0.613248721 0.139864034 0.102255667
763 SOLV HW1 2361 -0.550764707 0.106631246 0.031592642
763 SOLV HW2 2362 -0.564316588 0.147336614 0.189156037
764 SOLV OW 2363 1.383681905 0.313648858 1.349302608
764 SOLV HW1 2364 1.338071946 0.302351043 1.261020267
764 SOLV HW2 2365 1.482073877 0.299770210 1.337985110
765 SOLV OW 2366 1.288725279 0.190362611 0.555038237
765 SOLV HW1 2367 1.380833615 0.194496679 0.593756355
765 SOLV HW2 2368 1.292673091 0.149120087 0.464024177
766 SOLV OW 2369 -0.899230423 0.316671066 0.708075230
766 SOLV HW1 2370 -0.848593991 0.233927310 0.683759614
766 SOLV HW2 2371 -0.835439642 0.393048311 0.718027296
767 SOLV OW 2372 -0.567039685 1.238626892 -1.256318412
767 SOLV HW1 2373 -0.629197121 1.270661566 -1.184818029
767 SOLV HW2 2374 -0.548696319 1.141178562 -1.243303724
768 SOLV OW 2375 -0.337998685 -0.177562053 0.400661992
768 SOLV HW1 2376 -0.298754532 -0.241231258 0.467054123
768 SOLV HW2 2377 -0.393062289 -0.228304831 0.334367430
769 SOLV OW 2378 -0.105208194 0.531952216 1.324851566
769 SOLV HW1 2379 -0.152835065 0.565175664 1.243430718
769 SOLV HW2 2380 -0.160333181 0.551466216 1.405981120
770 SOLV OW 2381 -0.058688443 -0.987366043 1.119902095
770 SOLV HW1 2382 -0.119306427 -0.929970626 1.174959843
770 SOLV HW2 2383 -0.099478416 -1.078015394 1.108995063
771 SOLV OW 2384 0.929309697 -1.423888950 0.324276833
771 SOLV HW1 2385 1.014035109 -1.456623992 0.366129656
771 SOLV HW2 2386 0.855539102 -1.488665210 0.343344140
772 SOLV OW 2387 -0.816095292 1.474830488 -0.101674403
772 SOLV HW1 2388 -0.726649029 1.444269369 -0.134316625
772 SOLV HW2 2389 -0.843421384 1.419665034 -0.022869914
773 SOLV OW 2390 -0.948807341 -1.209127883 1.208567886
773 SOLV HW1 2391 -1.035185071 -1.252027344 1.234999196
773 SOLV HW2 2392 -0.918423046 -1.246626638 1.120973962
774 SOLV OW 2393 0.154999095 -1.500254263 -1.082550986
774 SOLV HW1 2394 0.234697602 -1.440485836 -1.073745610
774 SOLV HW2 2395 0.174606050 -1.588458609 -1.039688781
775 SOLV OW 2396 0.826631608 -0.846375168 -0.677853220
775 SOLV HW1 2397 0.814767178 -0.748404068 -0.661642672
775 SOLV HW2 2398 0.737165981 -0.888979138 -0.691366534
776 SOLV OW 2399 1.011290931 0.761539465 -0.993407654
776 SOLV HW1 2400 0.942723591 0.699636862 -1.031728927
776 SOLV HW2 2401 1.102579180 0.731105426 -1.020650835
777 SOLV OW 2402 1.478077342 -0.932352086 -0.107494383
777 SOLV HW1 2403 1.432946618 -0.891640119 -0.028074921
777 SOLV HW2 2404 1.447708880 -1.027038981 -0.118163921
778 SOLV OW 2405 -0.712819387 1.397659698 -0.914299188
778 SOLV HW1 2406 -0.684879529 1.307292152 -0.946778708
778 SOLV HW2 2407 -0.649637367 1.466720143 -0.949519053
779 SOLV OW 2408 -1.511088657 0.869410689 -0.578618568
779 SOLV HW1 2409 -1.427792837 0.910061068 -0.616161122
779 SOLV HW2 2410 -1.487631586 0.811563741 -0.500480885
780 SOLV OW 2411 0.329248585 -0.573252096 0.638865536
780 SOLV HW1 2412 0.366303926 -0.486594816 0.605414892
780 SOLV HW2 2413 0.399160810 -0.621071825 0.692035693
781 SOLV OW 2414 1.559042792 0.039276117 1.128904833
781 SOLV HW1 2415 1.461439089 0.051476885 1.110836844
781 SOLV HW2 2416 1.593978819 0.118887456 1.178333797
782 SOLV OW 2417 -1.307350648 0.130457510 0.596078850
782 SOLV HW1 2418 -1.289062839 0.174931087 0.508391013
782 SOLV HW2 2419 -1.367323317 0.051740114 0.581644583
783 SOLV OW 2420 0.951615824 1.469512112 1.375352417
783 SOLV HW1 2421 0.984986395 1.562080205 1.393220868
783 SOLV HW2 2422 1.028642847 1.410358799 1.351489187
784 SOLV OW 2423 -0.491938101 1.538221742 -1.070243984
784 SOLV HW1 2424 -0.424224809 1.469678579 -1.043441876
784 SOLV HW2 2425 -0.510016204 1.530624728 -1.168310636
785 SOLV OW 2426 0.913698999 0.536512347 0.349404554
785 SOLV HW1 2427 0.975296898 0.590250684 0.291786128
785 SOLV HW2 2428 0.821194054 0.574033674 0.343323744
786 SOLV OW 2429 -0.620175840 -1.231633481 0.553379844
786 SOLV HW1 2430 -0.642274259 -1.167567901 0.479832338
786 SOLV HW2 2431 -0.664413639 -1.319510783 0.535420218
787 SOLV OW 2432 0.853758195 1.393484404 0.496259724
787 SOLV HW1 2433 0.896623958 1.408262006 0.407129019
787 SOLV HW2 2434 0.764104050 1.351071449 0.483478262
788 SOLV OW 2435 1.114118346 1.235608427 -0.249257791
788 SOLV HW1 2436 1.202549300 1.196243358 -0.224118042
788 SOLV HW2 2437 1.097642491 1.317935426 -0.194922050
789 SOLV OW 2438 0.678213584 0.240794661 0.204132545
789 SOLV HW1 2439 0.611486009 0.192870087 0.261160669
789 SOLV HW2 2440 0.715474378 0.177580430 0.136182874
790 SOLV OW 2441 -0.280923463 -0.341615616 0.858104788
790 SOLV HW1 2442 -0.314554710 -0.327044686 0.951153809
790 SOLV HW2 2443 -0.194907531 -0.392528053 0.861358014
791 SOLV OW 2444 1.125405644 -0.176724586 0.577876378
791 SOLV HW1 2445 1.216129816 -0.185589201 0.618994151
791 SOLV HW2 2446 1.094322038 -0.266316519 0.546140201
792 SOLV OW 2447 -0.293088259 -1.459351305 -0.769632621
792 SOLV HW1 2448 -0.227236937 -1.386719367 -0.789373171
792 SOLV HW2 2449 -0.385963368 -1.422935575 -0.776680262
793 SOLV OW 2450 1.534203227 -0.671027163 1.462703701
793 SOLV HW1 2451 1.596701469 -0.609555205 1.414569879
793 SOLV HW2 2452 1.445853495 -0.672085533 1.415854643
794 SOLV OW 2453 -0.736394768 1.120067655 -1.040519402
794 SOLV HW1 2454 -0.831987025 1.134530426 -1.014930399
794 SOLV HW2 2455 -0.702626746 1.036438755 -0.997302403
795 SOLV OW 2456 -0.223471900 0.185488180 -0.757460156
795 SOLV HW1 2457 -0.288058016 0.223634848 -0.691326963
795 SOLV HW2 2458 -0.138394409 0.237985231 -0.755016712
796 SOLV OW 2459 -0.625067561 0.171405001 -0.775638038
796 SOLV HW1 2460 -0.653035215 0.168677739 -0.871618463
796 SOLV HW2 2461 -0.555885897 0.101107169 -0.759088325
797 SOLV OW 2462 0.718213259 -1.118813356 0.427596633
797 SOLV HW1 2463 0.809721071 -1.154805290 0.409354406
797 SOLV HW2 2464 0.724446747 -1.043739583 0.493375249
798 SOLV OW 2465 1.490634897 -1.125004918 0.477331455
798 SOLV HW1 2466 1.488790791 -1.155244082 0.572641069
798 SOLV HW2 2467 1.584579347 -1.102721791 0.451262466
799 SOLV OW 2468 -1.513497733 1.079038810 1.013123871
799 SOLV HW1 2469 -1.514374322 1.054212139 1.109997403
799 SOLV HW2 2470 -1.419291747 1.078939163 0.979557065
800 SOLV OW 2471 0.615947511 -1.041882534 -1.132519902
800 SOLV HW1 2472 0.705291534 -1.065199355 -1.094102299
800 SOLV HW2 2473 0.572237167 -1.124625406 -1.167803613
801 SOLV OW 2474 -1.241895820 1.321665707 0.111088464
801 SOLV HW1 2475 -1.195613878 1.388260719 0.052564620
801 SOLV HW2 2476 -1.309547853 1.368565488 0.167881362
802 SOLV OW 2477 0.019646436 1.052808051 0.623449363
802 SOLV HW1 2478 -0.014645497 1.145257936 0.640155786
802 SOLV HW2 2479 0.111431611 1.057935530 0.584065107
803 SOLV OW 2480 -0.583426800 -0.925743454 -0.592699968
803 SOLV HW1 2481 -0.591663684 -0.848249991 -0.655366133
803 SOLV HW2 2482 -0.521278622 -0.993675869 -0.631724076
804 SOLV OW 2483 -0.971826559 -1.197975877 0.378694635
804 SOLV HW1 2484 -0.877087251 -1.179789328 0.352323577
804 SOLV HW2 2485 -1.026450745 -1.115187943 0.365888667
805 SOLV OW 2486 -0.236170489 -0.871217239 -0.882668959
805 SOLV HW1 2487 -0.288228180 -0.905984412 -0.960662832
805 SOLV HW2 2488 -0.254939228 -0.927653236 -0.802267405
806 SOLV OW 2489 -0.121139466 0.026927315 -1.122233169
806 SOLV HW1 2490 -0.158438821 -0.036588422 -1.189882219
806 SOLV HW2 2491 -0.122838802 -0.015874145 -1.031862090
807 SOLV OW 2492 -0.847243827 -0.704831471 0.930777226
807 SOLV HW1 2493 -0.854009451 -0.694807984 1.030052153
807 SOLV HW2 2494 -0.792935416 -0.629665497 0.893328993
808 SOLV OW 2495 -1.073501574 1.242836499 0.346103263
808 SOLV HW1 2496 -0.998747201 1.308575501 0.355603662
808 SOLV HW2 2497 -1.145796369 1.282547889 0.289547745
809 SOLV OW 2498 -0.776618547 -1.548901872 -0.525095017
809 SOLV HW1 2499 -0.777870552 -1.449372319 -0.534800113
809 SOLV HW2 2500 -0.867494823 -1.580901972 -0.498272802
810 SOLV OW 2501 -0.858578104 -1.105064092 -0.208037589
810 SOLV HW1 2502 -0.823851767 -1.011449622 -0.213726801
810 SOLV HW2 2503 -0.925668997 -1.111272523 -0.134131299
811 SOLV OW 2504 -0.644615496 -1.179768412 0.835250965
811 SOLV HW1 2505 -0.550117258 -1.200072833 0.860928705
811 SOLV HW2 2506 -0.652718760 -1.179442321 0.735572742
812 SOLV OW 2507 -1.015973262 -0.742981724 0.445731848
812 SOLV HW1 2508 -1.005098995 -0.821872722 0.506228754
812 SOLV HW2 2509 -1.086441791 -0.762734028 0.377571192
813 SOLV OW 2510 -0.172489382 0.957872554 -0.414570762
813 SOLV HW1 2511 -0.172328732 0.935525272 -0.317090300
813 SOLV HW2 2512 -0.191761095 1.055294070 -0.426381755
814 SOLV OW 2513 -0.241750085 -0.331908244 0.575718502
814 SOLV HW1 2514 -0.241565826 -0.427118475 0.545113952
814 SOLV HW2 2515 -0.262825837 -0.328359500 0.673416254
815 SOLV OW 2516 -1.449298199 -1.285280610 0.313585319
815 SOLV HW1 2517 -1.539412972 -1.301306669 0.273303530
815 SOLV HW2 2518 -1.390487120 -1.364251601 0.296123079
816 SOLV OW 2519 1.470300524 -0.784256393 -1.226949593
816 SOLV HW1 2520 1.486214535 -0.879931928 -1.251341524
816 SOLV HW2 2521 1.400879519 -0.745514197 -1.287626681
817 SOLV OW 2522 -1.361226402 -1.182019666 -0.794823547
817 SOLV HW1 2523 -1.460670689 -1.173520826 -0.788599866
817 SOLV HW2 2524 -1.337881343 -1.249484794 -0.864848934
818 SOLV OW 2525 -0.395751236 0.829971904 0.762583139
818 SOLV HW1 2526 -0.350265707 0.904286816 0.811673726
818 SOLV HW2 2527 -0.330619020 0.786243647 0.700558373
819 SOLV OW 2528 0.371730614 -1.540390865 0.729593513
819 SOLV HW1 2529 0.402218936 -1.481812926 0.654486713
819 SOLV HW2 2530 0.274856604 -1.562173179 0.717636300
820 SOLV OW 2531 -0.671554827 0.995165816 -0.540437863
820 SOLV HW1 2532 -0.673156204 1.084538592 -0.585289946
820 SOLV HW2 2533 -0.698512075 0.924543606 -0.605917316
821 SOLV OW 2534 0.123992330 0.559646090 0.820102938
821 SOLV HW1 2535 0.141225402 0.598431442 0.910650206
821 SOLV HW2 2536 0.190233165 0.486979394 0.801834062
822 SOLV OW 2537 -1.135755168 0.343092502 -0.782877851
822 SOLV HW1 2538 -1.165956356 0.250504710 -0.760139669
822 SOLV HW2 2539 -1.215085076 0.403809793 -0.787539017
823 SOLV OW 2540 0.363405676 0.561730756 0.203359551
823 SOLV HW1 2541 0.391125624 0.654489408 0.228442853
823 SOLV HW2 2542 0.376457581 0.548484322 0.105095482
824 SOLV OW 2543 1.352027418 1.239812736 1.199063799
824 SOLV HW1 2544 1.390842382 1.231860248 1.290887810
824 SOLV HW2 2545 1.297571649 1.158428577 1.178748772
825 SOLV OW 2546 0.313331859 -1.142612035 -1.057649122
825 SOLV HW1 2547 0.261083639 -1.159505606 -1.141224609
825 SOLV HW2 2548 0.377693370 -1.217704092 -1.042854901
826 SOLV OW 2549 1.339130069 1.329255332 0.940528958
826 SOLV HW1 2550 1.408088252 1.386279199 0.895867002
826 SOLV HW2 2551 1.334262570 1.352753024 1.037615258
827 SOLV OW 2552 1.131226306 -0.235990352 -0.381120963
827 SOLV HW1 2553 1.116420244 -0.137088948 -0.380059135
827 SOLV HW2 2554 1.074469495 -0.277409158 -0.452289167
828 SOLV OW 2555 0.848054439 0.290160214 -1.305747391
828 SOLV HW1 2556 0.772621142 0.321623932 -1.363379931
828 SOLV HW2 2557 0.820032364 0.292861789 -1.209783971
829 SOLV OW 2558 -0.671236665 1.262407633 0.176593371
829 SOLV HW1 2559 -0.655461864 1.182538056 0.234663467
829 SOLV HW2 2560 -0.583580971 1.304652341 0.153529241
830 SOLV OW 2561 -0.339793890 -1.458276858 -0.493471833
830 SOLV HW1 2562 -0.334730322 -1.498679721 -0.584806706
830 SOLV HW2 2563 -0.435037642 -1.436844675 -0.471761686
831 SOLV OW 2564 -0.987768884 -0.637358661 0.148694144
831 SOLV HW1 2565 -1.046441872 -0.581464222 0.207303109
831 SOLV HW2 2566 -1.040143549 -0.668997219 0.069589241
832 SOLV OW 2567 -0.661707875 -1.491660060 1.104392207
832 SOLV HW1 2568 -0.619862777 -1.576654280 1.072350864
832 SOLV HW2 2569 -0.744768750 -1.512783532 1.155932137
833 SOLV OW 2570 -0.900775225 0.752414395 -1.157057484
833 SOLV HW1 2571 -0.829080583 0.813946621 -1.189855706
833 SOLV HW2 2572 -0.974726043 0.806098579 -1.116421297
834 SOLV OW 2573 0.125434668 1.001343462 -1.518244009
834 SOLV HW1 2574 0.153421223 1.076869429 -1.577525206
834 SOLV HW2 2575 0.195433601 0.929937301 -1.519969028
835 SOLV OW 2576 0.699825617 0.127297177 1.385170112
835 SOLV HW1 2577 0.705048151 0.153273546 1.481596476
835 SOLV HW2 2578 0.724847750 0.205580023 1.328181142
836 SOLV OW 2579 0.006251765 0.819059261 1.428242386
836 SOLV HW1 2580 0.003526529 0.721453435 1.449859234
836 SOLV HW2 2581 0.100138347 0.845747640 1.406455190
837 SOLV OW 2582 -0.867349668 -0.262190769 1.463619242
837 SOLV HW1 2583 -0.924869074 -0.216378795 1.395835877
837 SOLV HW2 2584 -0.802791536 -0.196413526 1.502443882
838 SOLV OW 2585 1.381967086 1.096535257 0.533705231
838 SOLV HW1 2586 1.432459628 1.055983592 0.609915926
838 SOLV HW2 2587 1.338638176 1.181466736 0.563890720
839 SOLV OW 2588 -1.267400447 -0.309572985 1.436174272
839 SOLV HW1 2589 -1.281824092 -0.241824181 1.508310586
839 SOLV HW2 2590 -1.261969427 -0.400812674 1.476762924
840 SOLV OW 2591 0.024713772 0.285172063 -0.746526571
840 SOLV HW1 2592 0.082932840 0.222689492 -0.798564368
840 SOLV HW2 2593 0.042591222 0.273760338 -0.648794122
841 SOLV OW 2594 0.802553040 1.430882840 -0.259688708
841 SOLV HW1 2595 0.897504127 1.416482576 -0.231779540
841 SOLV HW2 2596 0.778591575 1.527250545 -0.247812814
842 SOLV OW 2597 0.753935831 -0.126516911 1.320632541
842 SOLV HW1 2598 0.680427065 -0.182333750 1.282125903
842 SOLV HW2 2599 0.726224423 -0.030423744 1.320762106
843 SOLV OW 2600 1.265087165 -0.544232686 1.081503780
843 SOLV HW1 2601 1.166630991 -0.555348722 1.095082040
843 SOLV HW2 2602 1.290332686 -0.448767194 1.097334212
844 SOLV OW 2603 0.793067601 1.502499038 1.163425254
844 SOLV HW1 2604 0.858908762 1.503449750 1.238697592
844 SOLV HW2 2605 0.740677653 1.587687071 1.163916028
845 SOLV OW 2606 -0.800291853 -0.838173536 -0.122166821
845 SOLV HW1 2607 -0.803186644 -0.740005191 -0.103281539
845 SOLV HW2 2608 -0.852455478 -0.887096466 -0.052256617
846 SOLV OW 2609 -0.451792018 0.621688527 -0.083360447
846 SOLV HW1 2610 -0.364764936 0.572693682 -0.088629830
846 SOLV HW2 2611 -0.451595732 0.697098061 -0.149051409
847 SOLV OW 2612 -0.025706188 -0.460697340 -1.203091180
847 SOLV HW1 2613 0.050423805 -0.444134138 -1.140389549
847 SOLV HW2 2614 -0.110637946 -0.427782416 -1.161803031
848 SOLV OW 2615 -0.696745782 -0.055115520 -0.518263097
848 SOLV HW1 2616 -0.615584640 -0.068430810 -0.575160474
848 SOLV HW2 2617 -0.778815712 -0.063712462 -0.574763046
849 SOLV OW 2618 -1.092098504 -0.324556164 -1.267151566
849 SOLV HW1 2619 -1.012747461 -0.381550896 -1.245782827
849 SOLV HW2 2620 -1.098591660 -0.250187559 -1.200602619
850 SOLV OW 2621 -0.569908949 1.390347580 -0.196854771
850 SOLV HW1 2622 -0.496865865 1.436362153 -0.247344187
850 SOLV HW2 2623 -0.552411264 1.291893993 -0.195282078
851 SOLV OW 2624 0.250321240 0.509122244 -0.380069520
851 SOLV HW1 2625 0.342665606 0.497782386 -0.343408135
851 SOLV HW2 2626 0.238552302 0.603081980 -0.412212173
852 SOLV OW 2627 -1.352292809 0.842116093 0.291333093
852 SOLV HW1 2628 -1.331772553 0.930257123 0.248768036
852 SOLV HW2 2629 -1.360635097 0.772038440 0.220473060
853 SOLV OW 2630 0.998083032 1.161047342 1.062673744
853 SOLV HW1 2631 1.058194769 1.138511565 1.139357902
853 SOLV HW2 2632 0.978621417 1.259142133 1.063310866
854 SOLV OW 2633 0.341094743 1.132807439 -1.346931076
854 SOLV HW1 2634 0.397361810 1.050522551 -1.338879572
854 SOLV HW2 2635 0.271033670 1.118548562 -1.416856920
855 SOLV OW 2636 -0.238923153 -0.299961465 1.280827512
855 SOLV HW1 2637 -0.275265895 -0.266703874 1.367862172
855 SOLV HW2 2638 -0.176809073 -0.231654407 1.242384712
856 SOLV OW 2639 0.036976050 -1.078630024 0.695213579
856 SOLV HW1 2640 0.088719790 -1.137558686 0.633151335
856 SOLV HW2 2641 0.083229737 -1.074774411 0.783799263
857 SOLV OW 2642 1.367268140 -0.397907060 0.622780785
857 SOLV HW1 2643 1.366314208 -0.351814035 0.711528501
857 SOLV HW2 2644 1.419347662 -0.343088868 0.557326918
858 SOLV OW 2645 -0.586294276 1.497683473 -0.550723981
858 SOLV HW1 2646 -0.525251101 1.527179320 -0.477201875
858 SOLV HW2 2647 -0.548695112 1.526479187 -0.638808472
859 SOLV OW 2648 0.196248574 0.927170568 -0.637662479
859 SOLV HW1 2649 0.292520872 0.907569946 -0.656306295
859 SOLV HW2 2650 0.139759615 0.889447789 -0.711052394
860 SOLV OW 2651 -1.275649221 1.532482929 0.783649581
860 SOLV HW1 2652 -1.352966442 1.482192355 0.744988811
860 SOLV HW2 2653 -1.237786320 1.593816071 0.714321411
861 SOLV OW 2654 -1.185940400 0.047583443 0.885123483
861 SOLV HW1 2655 -1.209718330 0.104277207 0.964004163
861 SOLV HW2 2656 -1.121525156 -0.023501771 0.913394869
862 SOLV OW 2657 -0.483103583 0.770805521 1.523425401
862 SOLV HW1 2658 -0.502053775 0.831986185 1.446616226
862 SOLV HW2 2659 -0.456743641 0.824845647 1.603341490
863 SOLV OW 2660 -0.835641236 0.523020395 0.191103696
863 SOLV HW1 2661 -0.802500997 0.531792459 0.285053271
863 SOLV HW2 2662 -0.929885017 0.556061528 0.185800424
864 SOLV OW 2663 0.956347662 1.029076809 -1.031195771
864 SOLV HW1 2664 0.968854589 1.082002227 -0.947265923
864 SOLV HW2 2665 0.993042863 0.936946812 -1.018263512
865 SOLV OW 2666 -1.432413420 -0.510376185 0.103418901
865 SOLV HW1 2667 -1.515209272 -0.536783282 0.053927875
865 SOLV HW2 2668 -1.457581879 -0.471071853 0.191869513
866 SOLV OW 2669 1.162334062 -1.506770174 0.415976840
866 SOLV HW1 2670 1.152700666 -1.536648743 0.321031587
866 SOLV HW2 2671 1.239410657 -1.443482183 0.423319780
867 SOLV OW 2672 -0.647230032 0.807135414 0.638049563
867 SOLV HW1 2673 -0.638587368 0.779368458 0.542370983
867 SOLV HW2 2674 -0.556275828 0.814370810 0.678977738
868 SOLV OW 2675 -1.161799480 -0.873546168 -1.045713973
868 SOLV HW1 2676 -1.204763831 -0.927323623 -1.118254843
868 SOLV HW2 2677 -1.131548218 -0.785745049 -1.082807297
869 SOLV OW 2678 -1.000765346 -1.262610616 -0.778838370
869 SOLV HW1 2679 -1.004690633 -1.286614386 -0.875843825
869 SOLV HW2 2680 -1.048441237 -1.332346352 -0.725307199
870 SOLV OW 2681 -0.279098693 0.685126257 0.580606958
870 SOLV HW1 2682 -0.300589539 0.602260162 0.528906486
870 SOLV HW2 2683 -0.235914824 0.751942460 0.520007362
871 SOLV OW 2684 0.717390483 0.271754274 -1.063898505
871 SOLV HW1 2685 0.649689396 0.212206019 -1.020626720
871 SOLV HW2 2686 0.802996266 0.267839957 -1.012339829
872 SOLV OW 2687 -0.978150105 1.235733222 1.217246261
872 SOLV HW1 2688 -1.056415860 1.289574282 1.248510204
872 SOLV HW2 2689 -0.955438013 1.261422207 1.123299426
873 SOLV OW 2690 0.979523343 -0.626160301 -1.121063310
873 SOLV HW1 2691 0.959193707 -0.713745932 -1.164849243
873 SOLV HW2 2692 1.071180237 -0.629333905 -1.081180667
874 SOLV OW 2693 0.686870693 -0.937614640 0.209753468
874 SOLV HW1 2694 0.639250230 -0.854177575 0.237546074
874 SOLV HW2 2695 0.699467014 -0.996862078 0.289332902
875 SOLV OW 2696 -1.368602050 1.290313705 1.201698146
875 SOLV HW1 2697 -1.363426586 1.191160691 1.213615508
875 SOLV HW2 2698 -1.333107613 1.314918013 1.111504670
876 SOLV OW 2699 -0.206168054 -1.418921211 0.906767227
876 SOLV HW1 2700 -0.264731900 -1.400402291 0.827843169
876 SOLV HW2 2701 -0.188845575 -1.517215907 0.913064344
877 SOLV OW 2702 0.370051201 -0.131815238 1.360261912
877 SOLV HW1 2703 0.379038501 -0.032228013 1.362176154
877 SOLV HW2 2704 0.273285622 -0.156626095 1.365048436
878 SOLV OW 2705 1.427658978 0.262339099 0.910827511
878 SOLV HW1 2706 1.388060758 0.171788019 0.926127508
878 SOLV HW2 2707 1.470998569 0.264959138 0.820736493
879 SOLV OW 2708 0.594111290 1.404680752 0.806231841
879 SOLV HW1 2709 0.667387803 1.357401697 0.757289694
879 SOLV HW2 2710 0.572950936 1.355125121 0.890484200
880 SOLV OW 2711 0.869504683 -1.418069318 0.059965789
880 SOLV HW1 2712 0.892837984 -1.438732698 0.154994337
880 SOLV HW2 2713 0.806071114 -1.340798620 0.057284406
881 SOLV OW 2714 1.054204647 0.129972121 -1.357841750
881 SOLV HW1 2715 1.126503651 0.193024472 -1.386108269
881 SOLV HW2 2716 0.982948199 0.180614508 -1.309266686
882 SOLV OW 2717 0.075486573 0.683606423 0.573627502
882 SOLV HW1 2718 0.004864581 0.753220329 0.586599437
882 SOLV HW2 2719 0.106510786 0.650606552 0.662791732
883 SOLV OW 2720 -0.848145734 -1.323037097 0.966636773
883 SOLV HW1 2721 -0.792987459 -1.255284064 0.917968162
883 SOLV HW2 2722 -0.789697558 -1.398668688 0.996050361
884 SOLV OW 2723 -0.104816648 0.942258806 -0.125171106
884 SOLV HW1 2724 -0.011384323 0.906785025 -0.128878559
884 SOLV HW2 2725 -0.160406914 0.883605294 -0.066254512
885 SOLV OW 2726 1.343486189 -0.484130060 -0.010936555
885 SOLV HW1 2727 1.322624531 -0.492109424 0.086544671
885 SOLV HW2 2728 1.300189031 -0.559402016 -0.060544279
886 SOLV OW 2729 -0.669453066 -1.426819094 1.491946702
886 SOLV HW1 2730 -0.573645476 -1.436164782 1.464824585
886 SOLV HW2 2731 -0.702845505 -1.336297306 1.465626930
887 SOLV OW 2732 -0.397784488 -0.552923258 -0.899395196
887 SOLV HW1 2733 -0.403076087 -0.564158263 -0.998630933
887 SOLV HW2 2734 -0.315822354 -0.598765180 -0.865004771
888 SOLV OW 2735 -0.837244631 -0.449783956 -0.893448779
888 SOLV HW1 2736 -0.772336451 -0.412713946 -0.959892134
888 SOLV HW2 2737 -0.787809084 -0.480307403 -0.812045842
889 SOLV OW 2738 -0.629303922 -0.960204641 -1.368969199
889 SOLV HW1 2739 -0.659278887 -0.931757510 -1.277906644
889 SOLV HW2 2740 -0.625728291 -0.880506666 -1.429264619
890 SOLV OW 2741 -0.988577143 0.448456405 -0.977771165
890 SOLV HW1 2742 -0.924368362 0.393024616 -1.030744646
890 SOLV HW2 2743 -1.033472544 0.391083135 -0.909256738
891 SOLV OW 2744 0.331501016 1.255444608 0.654733284
891 SOLV HW1 2745 0.370836171 1.193643381 0.586651768
891 SOLV HW2 2746 0.359607392 1.349309054 0.634702958
892 SOLV OW 2747 -0.588050114 -0.921079586 0.503957549
892 SOLV HW1 2748 -0.538759642 -0.934753407 0.418020535
892 SOLV HW2 2749 -0.536325620 -0.962326356 0.578957205
893 SOLV OW 2750 0.507085817 0.282274440 -0.684725079
893 SOLV HW1 2751 0.487437493 0.267643005 -0.781688347
893 SOLV HW2 2752 0.454097965 0.217582459 -0.629868646
894 SOLV OW 2753 1.340900003 -1.552009603 -0.964650434
894 SOLV HW1 2754 1.439988684 -1.542293697 -0.955229625
894 SOLV HW2 2755 1.303283823 -1.469811917 -1.007430931
895 SOLV OW 2756 -0.669156400 -1.373521569 -1.097416423
895 SOLV HW1 2757 -0.661755104 -1.470015032 -1.072195930
895 SOLV HW2 2758 -0.618795142 -1.357314261 -1.182285811
896 SOLV OW 2759 1.098415037 0.036605178 -0.392538363
896 SOLV HW1 2760 1.158934520 0.089732406 -0.333237218
896 SOLV HW2 2761 1.041119925 0.098782399 -0.445952003
897 SOLV OW 2762 -0.703039036 0.064631724 1.303525114
897 SOLV HW1 2763 -0.607243597 0.072984884 1.276050027
897 SOLV HW2 2764 -0.757468631 0.033585920 1.225582566
898 SOLV OW 2765 1.162013356 -1.052228036 -0.705971196
898 SOLV HW1 2766 1.158035016 -1.021586696 -0.610854345
898 SOLV HW2 2767 1.236578838 -1.005073617 -0.753068937
899 SOLV OW 2768 -1.549680385 -1.186585060 1.087555538
899 SOLV HW1 2769 -1.595482316 -1.103506994 1.119209433
899 SOLV HW2 2770 -1.455811446 -1.164756737 1.060837171
900 SOLV OW 2771 1.266816290 0.978023374 1.110036268
900 SOLV HW1 2772 1.335513798 0.983582276 1.037570773
900 SOLV HW2 2773 1.273264249 0.889082476 1.155306007
901 SOLV OW 2774 0.847140038 -0.539277572 -1.388684432
901 SOLV HW1 2775 0.792011243 -0.544779208 -1.305422609
901 SOLV HW2 2776 0.899058165 -0.624013753 -1.399919421
902 SOLV OW 2777 -1.238703148 0.975705858 1.022392149
902 SOLV HW1 2778 -1.244962399 0.879281339 1.048146074
902 SOLV HW2 2779 -1.142784519 1.000286102 1.008409360
903 SOLV OW 2780 1.048019896 -1.329605541 -0.134566502
903 SOLV HW1 2781 1.093554585 -1.397678608 -0.191965919
903 SOLV HW2 2782 0.995537755 -1.375974248 -0.063169857
904 SOLV OW 2783 -1.420155460 -0.840102131 -0.574077182
904 SOLV HW1 2784 -1.511901647 -0.820891556 -0.539217476
904 SOLV HW2 2785 -1.372361791 -0.753966124 -0.591336753
905 SOLV OW 2786 1.464561608 1.223449705 -1.408584189
905 SOLV HW1 2787 1.380565399 1.275965909 -1.394858857
905 SOLV HW2 2788 1.541188628 1.286197451 -1.422468595
906 SOLV OW 2789 -0.318189005 -1.342240999 -1.106552780
906 SOLV HW1 2790 -0.269749052 -1.428751180 -1.119630315
906 SOLV HW2 2791 -0.400458595 -1.341197256 -1.163403931
907 SOLV OW 2792 -0.495399763 1.512795296 1.450248740
907 SOLV HW1 2793 -0.436673788 1.450882883 1.502385500
907 SOLV HW2 2794 -0.581270150 1.466547797 1.428168533
908 SOLV OW 2795 1.129248972 0.586196048 -1.390764510
908 SOLV HW1 2796 1.112420428 0.657904904 -1.458412391
908 SOLV HW2 2797 1.075034976 0.505354139 -1.413720156
909 SOLV OW 2798 -0.544390349 0.194067404 0.818206597
909 SOLV HW1 2799 -0.465562206 0.247735457 0.788080801
909 SOLV HW2 2800 -0.578185085 0.138840217 0.741986140
910 SOLV OW 2801 1.456441403 -0.189222488 -0.543556238
910 SOLV HW1 2802 1.377396140 -0.135060574 -0.572198180
910 SOLV HW2 2803 1.477384517 -0.257294856 -0.613766306
911 SOLV OW 2804 -0.660244748 0.818360677 0.051144775
911 SOLV HW1 2805 -0.725106595 0.884033625 0.012654858
911 SOLV HW2 2806 -0.606149485 0.777848945 -0.022570467
912 SOLV OW 2807 -0.190024460 0.822191631 1.214547420
912 SOLV HW1 2808 -0.149005566 0.905579800 1.177590746
912 SOLV HW2 2809 -0.147400956 0.800162708 1.302296262
913 SOLV OW 2810 0.132118937 1.244412497 1.467928375
913 SOLV HW1 2811 0.130743899 1.144440787 1.470225172
913 SOLV HW2 2812 0.039823280 1.278603278 1.450208407
914 SOLV OW 2813 1.440693331 1.080680547 -0.250508634
914 SOLV HW1 2814 1.458853168 1.149343158 -0.320906060
914 SOLV HW2 2815 1.419085636 0.993274831 -0.294022294
915 SOLV OW 2816 0.822699638 0.002831804 -1.474632211
915 SOLV HW1 2817 0.795736132 0.095403858 -1.501190527
915 SOLV HW2 2818 0.918565473 0.002940513 -1.446142486
916 SOLV OW 2819 0.474617473 -1.007014000 -0.448397199
916 SOLV HW1 2820 0.396352258 -1.038038348 -0.394417252
916 SOLV HW2 2821 0.551334593 -0.987064762 -0.387419178
917 SOLV OW 2822 -1.015220788 -1.335595873 -1.036942121
917 SOLV HW1 2823 -0.950661507 -1.298549965 -1.103738066
917 SOLV HW2 2824 -0.994380729 -1.432000302 -1.020392753
918 SOLV OW 2825 -0.468894177 -0.099236082 -0.652970832
918 SOLV HW1 2826 -0.409215546 -0.030397859 -0.611719351
918 SOLV HW2 2827 -0.415064462 -0.180144485 -0.676592630
919 SOLV OW 2828 1.344530393 -0.820823275 -0.809738777
919 SOLV HW1 2829 1.361620778 -0.912061772 -0.846935347
919 SOLV HW2 2830 1.414775120 -0.757730503 -0.842679186
920 SOLV OW 2831 -1.488082410 0.486839369 0.201325392
920 SOLV HW1 2832 -1.474699858 0.550788298 0.125608833
920 SOLV HW2 2833 -1.529208549 0.402468877 0.166802955
921 SOLV OW 2834 -0.889294672 0.806924744 -0.595201754
921 SOLV HW1 2835 -0.885539177 0.897474874 -0.552914740
921 SOLV HW2 2836 -0.842868808 0.740721165 -0.536352191
922 SOLV OW 2837 1.200711992 1.511415568 0.588170438
922 SOLV HW1 2838 1.156851190 1.601259047 0.590685720
922 SOLV HW2 2839 1.187943450 1.470632528 0.497751401
923 SOLV OW 2840 1.287996029 0.667307297 -0.827983161
923 SOLV HW1 2841 1.271826602 0.647173224 -0.924600991
923 SOLV HW2 2842 1.381506665 0.641413808 -0.803754324
924 SOLV OW 2843 -0.319431397 0.489121414 -1.048463168
924 SOLV HW1 2844 -0.357399890 0.531506277 -0.966220522
924 SOLV HW2 2845 -0.219616816 0.488016538 -1.042320923
925 SOLV OW 2846 -0.973679729 0.656386950 0.489735498
925 SOLV HW1 2847 -1.054567821 0.694468742 0.444915317
925 SOLV HW2 2848 -0.949689722 0.712832730 0.568730310
926 SOLV OW 2849 0.607828465 0.878953327 0.671274174
926 SOLV HW1 2850 0.588639251 0.950188923 0.738782652
926 SOLV HW2 2851 0.696751258 0.837604021 0.690846143
927 SOLV OW 2852 -0.138582654 1.209490662 -0.907010207
927 SOLV HW1 2853 -0.080143612 1.273195927 -0.957275965
927 SOLV HW2 2854 -0.088086967 1.172373825 -0.829083215
928 SOLV OW 2855 1.426922275 -1.412550665 1.511924855
928 SOLV HW1 2856 1.433233754 -1.458405281 1.423272475
928 SOLV HW2 2857 1.492988986 -1.453039025 1.575151767
929 SOLV OW 2858 0.437957497 0.846891043 -0.739410017
929 SOLV HW1 2859 0.475387172 0.773789595 -0.796481114
929 SOLV HW2 2860 0.512708523 0.903158876 -0.704082468
930 SOLV OW 2861 0.170534368 0.497858148 1.303330937
930 SOLV HW1 2862 0.222284399 0.525786064 1.384223680
930 SOLV HW2 2863 0.072652580 0.501419878 1.323532562
END
GENBOX
1
3.077767162 3.077767162 3.077767162
90.000000000 90.000000000 90.000000000
0.000000000 -0.000000000 0.000000000
0.000000000 0.000000000 0.000000000
END
Perform Emin¶
Now we will perform the solvent box energy minimization with the restrained peptide.
[47]:
# run emin
out_eminBox_system = emin(in_gromos_system=in_eminBox_system,
step_name="e_"+in_eminBox_system.name)
################################################################################
e_emin_solvBox
################################################################################
Script: /home/mlehner/PyGromosTools/pygromos/simulations/hpc_queuing/job_scheduling/schedulers/simulation_scheduler.py
################################################################################
Simulation Setup:
################################################################################
steps_per_run: 3000
equis: 0
simulation runs: 1
################################################################################
submit final analysis part
/home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/e_emin_solvBox/ana_out.log
/home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/e_emin_solvBox/job_analysis.py
ANA jobID: 0
Whoops what happened here? What you see is the effect of the periodic box and the peptide placed directly at the corners of the box. This is absolutely ok and there is nothing to worry about. We can recenter the peptide and then we see the expected same configuration like from the vacuum emin.
[48]:
#show resulting coordinates
view = out_eminBox_system.cnf.view
view
Now lets fix that!
[49]:
#out_eminBox_system.cnf.recenter_pbc()
out_eminBox_system.cnf.recreate_view()
recentered_view = out_eminBox_system.cnf.view
recentered_view
[50]:
out_eminBox_system_path = out_eminBox_system.save(out_eminBox_system.work_folder+"/emin_box_result.obj")
Thermalisation and equilibration.¶
In the previous steps you have generated a topology and initial coordinates of your system. At this point, you have to generate initial velocities. In the process of thermalisation and equilibration, initial velocities are sampled from a Maxwell-Boltzmann distribution at a low temperature and the system is slowly heated up to the final production simulation temperature. The atoms of the solute are again positionally restrained and these restraints are loosened while heating up. With the help of these restraints you make sure that the random initial velocities do not disrupt the initial conformation too much.
Simulation Paramters¶
To performe the described equilibration we require additional Simulation Parameter blocks that will be described in the following. First we will load a simulation parameter template for MD simulations.
[51]:
from pygromos.data.simulation_parameters_templates import template_md_tut as template_md
from pygromos.files.simulation_parameters import imd
md_imd_file = imd.Imd(template_md) #in_eq_system.imd
Initialise Block¶
In the INITIALISE block the NTIVEL tells GROMOS whether it should generate the initial velocities or read them from the configuration file. NTISHK is used to restore bond length constraints (SHAKE). NTINHT and NTINHB are only used for Nose-Hoover thermo- and barostats and can be ignored in our case. Every time an atom is leaving the periodic box and entering it from the opposite site this incident is recorded in the so-called lattice shift vectors. Using NTISHI we want to make sure that these vectors are initialised to zero. As you don’t want to use roto-translational constraints NTIRTC can be ignored. NTICOM is used for initial removal of centre of mass motion. NTISTI is used to reset the stochastic integrals used in stochastic dynamics (SD) simulations. IG is the random number generator seed and TEMPI the initial temperature used to generate the Maxwell-Boltzmann distribution for generation of initial velocities. See also 4-94 for more information.
[52]:
help(md_imd_file.INITIALISE)
Help on INITIALISE in module pygromos.files.blocks.imd_blocks object:
class INITIALISE(_generic_imd_block)
| INITIALISE(NTIVEL: bool = False, NTISHK: int = 0, NTINHT: bool = False, NTINHB: bool = False, NTISHI: bool = False, NTIRTC: bool = False, NTICOM: int = 0, NTISTI: bool = False, IG: int = 0, TEMPI: float = 0, content: List[str] = None)
|
| INITIALISE block
|
| This block controls the Initialisation of a simulation.
|
| Attributes
| ----------
| NTIVEL: bool
| 0,1 controls generation of initial velocities
| 0: read from configuration (default)
| 1: generate from Maxell distribution at temperature TEMPI
| NTISHK: int
| 0..3 controls shaking of initial configuration
| 0: no intial SHAKE (default)
| 1: initial SHAKE on coordinates only
| 2: initial SHAKE on velocities only
| 3: initial SHAKE on coordinates and velocities
| NTINHT: bool
| 0,1 controls generation of initial Nose-Hoover chain variables
| 0: read from configuration (default)
| 1: reset variables to zero.
| NTINHB: bool
| 0,1 controls generation of initial Nose-Hoover (chain) barostat variables
| 0: read from strartup file (if applicable) (default)
| 1: reset variables to zero
| NTISHI: bool
| 0,1 controls initial setting for lattice shift vectors
| 0: read from configuration (default)
| 1: reset shifts to zero.
| NTIRTC: bool
| 0,1 controls initial setting of positions and orientations for roto-translational constraints
| 0: read from configuration (default)
| 1: reset based on initial configuraion of startup file
| NTICOM: int
| 0,1,2 controls initial removal of COM motion
| 0: no initial system COM motion removal (default)
| 1: initial COM translation is removed
| 2: initial COM rotation is removed
| NTISTI: bool
| 0,1 controls generation of stochastic integrals
| 0: read stochastic integrals and IG from configuration (default)
| 1: set stochastic integrals to zero and use IG from here.
| IG: int
| random number generator seed
| TEMPI: float
| initial temperature
|
| Method resolution order:
| INITIALISE
| _generic_imd_block
| pygromos.files.blocks._general_blocks._generic_gromos_block
| builtins.object
|
| Methods defined here:
|
| __init__(self, NTIVEL: bool = False, NTISHK: int = 0, NTINHT: bool = False, NTINHB: bool = False, NTISHI: bool = False, NTIRTC: bool = False, NTICOM: int = 0, NTISTI: bool = False, IG: int = 0, TEMPI: float = 0, content: List[str] = None)
| Args:
| NTIVEL:
| NTISHK:
| NTINHT:
| NTINHB:
| NTISHI:
| NTIRTC:
| NTICOM:
| NTISTI:
| IG:
| TEMPI:
|
| ----------------------------------------------------------------------
| Data and other attributes defined here:
|
| __annotations__ = {'IG': <class 'int'>, 'NTICOM': <class 'int'>, 'NTIN...
|
| __slotnames__ = []
|
| name = 'INITIALISE'
|
| ----------------------------------------------------------------------
| Methods inherited from _generic_imd_block:
|
| block_to_string(self) -> str
|
| read_content_from_str(self, content: List[str])
|
| ----------------------------------------------------------------------
| Methods inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __copy__(self)
|
| __deepcopy__(self, memo)
|
| __eq__(self, _generic_gromos_block__o: object) -> bool
| Return self==value.
|
| __iter__(self)
|
| __repr__(self)
| Return repr(self).
|
| __str__(self)
| Return str(self).
|
| get_name(self)
|
| ----------------------------------------------------------------------
| Data descriptors inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
|
| ----------------------------------------------------------------------
| Data and other attributes inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __hash__ = None
And now we will check the set options in our template:
[53]:
md_imd_file.INITIALISE
[53]:
INITIALISE
# NTIVEL NTISHK NTINHT NTINHB
1 0 0 0
# NTISHI NTIRTC NTICOM
1 0 1
# NTISTI
0
# IG TEMPI
12416 300.000000
END
System Block¶
In the SYSTEM block you would need to replace NSM with the number of solvent molecules in your system, but the prepare_simulation function takes care of that for you.
[54]:
#The last SOLV molecule can be found out like this:
str(out_eminBox_system.cnf.POSITION[-1])
[54]:
' 930 SOLV HW2 2863 0.186473330 0.337573696 1.044135675\n'
[55]:
#the template currently contains these values - note NSM is 1172, which is wrong!
md_imd_file.SYSTEM
[55]:
SYSTEM
# NPM NSM
1 5
END
STEP Block¶
In the STEP block you specify how many steps you want to simulate (NSTLIM), at what time your simulation starts (T) and how big the integration time step (DT) is. In this case you want to start at time 0 and you want to carry out a 20 ps simulation, because the time unit happens to be ps.
[56]:
help(md_imd_file.STEP)
Help on STEP in module pygromos.files.blocks.imd_blocks object:
class STEP(_generic_imd_block)
| STEP(NSTLIM: int = 0, T: float = 0, DT: float = 0, content: List[str] = None)
|
| Step Block
|
| This Block gives the number of simulation steps,
|
| Attributes
| -----------
| NSTLIM: int
| number of simulations Step till terminating.
| T: float
| Starting Time
| DT: float
| time step [fs]
|
| Method resolution order:
| STEP
| _generic_imd_block
| pygromos.files.blocks._general_blocks._generic_gromos_block
| builtins.object
|
| Methods defined here:
|
| __init__(self, NSTLIM: int = 0, T: float = 0, DT: float = 0, content: List[str] = None)
| Initialize self. See help(type(self)) for accurate signature.
|
| ----------------------------------------------------------------------
| Data and other attributes defined here:
|
| __annotations__ = {'DT': <class 'float'>, 'NSTLIM': <class 'int'>, 'T'...
|
| __slotnames__ = []
|
| name = 'STEP'
|
| ----------------------------------------------------------------------
| Methods inherited from _generic_imd_block:
|
| block_to_string(self) -> str
|
| read_content_from_str(self, content: List[str])
|
| ----------------------------------------------------------------------
| Methods inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __copy__(self)
|
| __deepcopy__(self, memo)
|
| __eq__(self, _generic_gromos_block__o: object) -> bool
| Return self==value.
|
| __iter__(self)
|
| __repr__(self)
| Return repr(self).
|
| __str__(self)
| Return str(self).
|
| get_name(self)
|
| ----------------------------------------------------------------------
| Data descriptors inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
|
| ----------------------------------------------------------------------
| Data and other attributes inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __hash__ = None
[57]:
md_imd_file.STEP
[57]:
STEP
# NSTLIM T DT
1000 0.000000 0.002000
END
BOUNDCOND Block¶
As previously described, with the BOUNDCOND block you specify which PBC you will use. With NTB=1 you specify rectangular PBC.
[58]:
help(md_imd_file.BOUNDCOND)
Help on BOUNDCOND in module pygromos.files.blocks.imd_blocks object:
class BOUNDCOND(_generic_imd_block)
| BOUNDCOND(NTB: int = 0, NDFMIN: int = 0, content: List[str] = None)
|
| Boundary Condition Block
|
| This block describes the boundary condition of the coordinate system.
|
| Attributes
| ----------
| NTB : int, optional
| Boundary conditions, by default 0
| -1 : truncated octahedral
| 0 : vacuum
| 1 : rectangular
| 2 : triclinic
| NDFMIN : int, optional
| number of degrees of freedom subtracted for temperature, by default 0
|
| Method resolution order:
| BOUNDCOND
| _generic_imd_block
| pygromos.files.blocks._general_blocks._generic_gromos_block
| builtins.object
|
| Methods defined here:
|
| __init__(self, NTB: int = 0, NDFMIN: int = 0, content: List[str] = None)
| Initialize self. See help(type(self)) for accurate signature.
|
| ----------------------------------------------------------------------
| Data and other attributes defined here:
|
| __annotations__ = {'NDFMIN': <class 'int'>, 'NTB': <class 'int'>, 'nam...
|
| __slotnames__ = []
|
| name = 'BOUNDCOND'
|
| ----------------------------------------------------------------------
| Methods inherited from _generic_imd_block:
|
| block_to_string(self) -> str
|
| read_content_from_str(self, content: List[str])
|
| ----------------------------------------------------------------------
| Methods inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __copy__(self)
|
| __deepcopy__(self, memo)
|
| __eq__(self, _generic_gromos_block__o: object) -> bool
| Return self==value.
|
| __iter__(self)
|
| __repr__(self)
| Return repr(self).
|
| __str__(self)
| Return str(self).
|
| get_name(self)
|
| ----------------------------------------------------------------------
| Data descriptors inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
|
| ----------------------------------------------------------------------
| Data and other attributes inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __hash__ = None
[59]:
md_imd_file.BOUNDCOND
[59]:
BOUNDCOND
# NTB NDFMIN
1 3
END
MULTIBATH Block¶
In our case we want to run the simulation at constant temperature. For this purpose, we have to add the MULTIBATH input block (see 4-96). First we specify which algorithm we will use. In this case we will use the weak-coupling scheme (ALGORITHM=0). How many temperature baths we want to couple to the system is specified by NBATHS. You can specify the temperature using the TEMP0 parameter. TAUT is the coupling time used in the weak-coupling method for this bath. DOFSET specifies the number of distiguishable sets of degrees of freedom. LAST is pointing to the last atom for the set of degrees of freedom; thus, you put the number of last atom of your system instead of LSTATM. COMBATH is the temperature bath to which we want to couple the centre of mass motion of this set of degrees of freedom IRBATH is the temperature bath to which the internal and rotational degrees of freedom of this set of degrees of freedom are coupled.
[60]:
help(md_imd_file.MULTIBATH)
Help on MULTIBATH in module pygromos.files.blocks.imd_blocks object:
class MULTIBATH(_generic_imd_block)
| MULTIBATH(ALGORITHM: int = 0, NBATHS: int = 0, TEMP0: List[float] = [], TAU: List[float] = [], DOFSET: int = 0, LAST: List[int] = [], COMBATH: List[int] = [], IRBATH: List[int] = [], NUM: int = None, content: List[str] = None)
|
| MULTIBATH Block
|
| This block controls the Thermostat of a simulation. Multiple temperature baths can be coupled.
|
| Attributes
| ----------
| ALGORITHM: int
| temperature coupling algorithm
| weak-coupling(0)
| nose-hoover(1)
| nose-hoover-chains(2) num
| (where num is the number of chains to use)
| NUM: int, optional
| Mumber of chains in Nosé Hoover chains scheme [only specify when needed]
| NBATHS: int
| Number of temperature baths
| TEMP0: List[float]
| temperature of each bath (list len == NBATHS)
| TAU: List[float]
| coupling of the temperaturebath to the system (list len == NBATHS)
| DOFSET: int
| Number of set of Degrees of freedom.
| LAST: List[int] (list len == DOFSET)
| Last atoms of each DOFSet
| COMBATH: List[int] (list len == DOFSET)
| Index of the temperature baths
| IRBATH: List[int] (list len == DOFSET)
| IRBAHT?
|
| See Also
| --------
| _generic_imd_block, _generic_gromos_block
|
| Method resolution order:
| MULTIBATH
| _generic_imd_block
| pygromos.files.blocks._general_blocks._generic_gromos_block
| builtins.object
|
| Methods defined here:
|
| __init__(self, ALGORITHM: int = 0, NBATHS: int = 0, TEMP0: List[float] = [], TAU: List[float] = [], DOFSET: int = 0, LAST: List[int] = [], COMBATH: List[int] = [], IRBATH: List[int] = [], NUM: int = None, content: List[str] = None)
| Initialize self. See help(type(self)) for accurate signature.
|
| adapt_multibath(self, last_atoms_bath: Dict[int, int], algorithm: int = None, num: int = None, T: Union[float, List[float]] = None, TAU: float = None) -> None
| adapt_multibath
| This function is adding each atom set into a single multibath.
| #TODO implementation not correct with com_bath and irbath! Works for super simple cases though
|
| Parameters
| ----------
| last_atoms_bath : Dict[int,int]
| lastatom:bath
| algorithm : int
| int code for algorihtm
| T : float,List[float], optional
| temperature value
| TAU : float, optional
| coupling var
|
| Returns
| -------
| None
|
| block_to_string(self) -> str
|
| read_content_from_str(self, content: List[str])
|
| ----------------------------------------------------------------------
| Data and other attributes defined here:
|
| __annotations__ = {'ALGORITHM': <class 'int'>, 'COMBATH': typing.List[...
|
| name = 'MULTIBATH'
|
| ----------------------------------------------------------------------
| Methods inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __copy__(self)
|
| __deepcopy__(self, memo)
|
| __eq__(self, _generic_gromos_block__o: object) -> bool
| Return self==value.
|
| __iter__(self)
|
| __repr__(self)
| Return repr(self).
|
| __str__(self)
| Return str(self).
|
| get_name(self)
|
| ----------------------------------------------------------------------
| Data descriptors inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
|
| ----------------------------------------------------------------------
| Data and other attributes inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __hash__ = None
[61]:
md_imd_file.MULTIBATH
[61]:
MULTIBATH
# ALGORITHM
0
# NBATHS
2
# TEMP0(1 ... NBATHS) TAU(1 ... NBATHS)
300.0 0.1
300.0 0.1
# DOFSET
2
# LAST(1 ... DOFSET) COMBATH(1 ... DOFSET) IRBATH(1 ... DOFSET)
73 1 1
2863 2 2
END
COMTRANSROT Block¶
This block is needed to remove the centre of mass motion (translation and rotation). Without this block it can happen that all the kinetic energy is converted to centre of mass translation (flying ice cube problem). With NSCM we specify how often the center-of-mass (COM) motion is removed. If NSCM is \(< 0\) translation and rotation motion are removed every \(|\) NSCM \(|\) th step. If NSCM is \(> 0\) only translation motion is removed every NSCMth step.
[62]:
help(md_imd_file.COMTRANSROT)
Help on COMTRANSROT in module pygromos.files.blocks.imd_blocks object:
class COMTRANSROT(_generic_imd_block)
| COMTRANSROT(NSCM: int = 0, content: List[str] = None)
|
| COMTRANSROT block
|
| This block controls the center of mass translation and rotation removal. (flying ice cube problem)
|
| Attributes
| ----------
|
| NSCM : int
| controls system centre-of-mass (com) motion removal
| 0: no com motion removal (default)
| < 0: com translation and rotation are removed every abs(NSCM) steps.
| > 0: com translation is removed every NSCM steps.
|
| Method resolution order:
| COMTRANSROT
| _generic_imd_block
| pygromos.files.blocks._general_blocks._generic_gromos_block
| builtins.object
|
| Methods defined here:
|
| __init__(self, NSCM: int = 0, content: List[str] = None)
| Initialize self. See help(type(self)) for accurate signature.
|
| ----------------------------------------------------------------------
| Data and other attributes defined here:
|
| __annotations__ = {'NSCM': <class 'int'>, 'name': <class 'str'>}
|
| name = 'COMTRANSROT'
|
| ----------------------------------------------------------------------
| Methods inherited from _generic_imd_block:
|
| block_to_string(self) -> str
|
| read_content_from_str(self, content: List[str])
|
| ----------------------------------------------------------------------
| Methods inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __copy__(self)
|
| __deepcopy__(self, memo)
|
| __eq__(self, _generic_gromos_block__o: object) -> bool
| Return self==value.
|
| __iter__(self)
|
| __repr__(self)
| Return repr(self).
|
| __str__(self)
| Return str(self).
|
| get_name(self)
|
| ----------------------------------------------------------------------
| Data descriptors inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
|
| ----------------------------------------------------------------------
| Data and other attributes inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __hash__ = None
[63]:
md_imd_file.COMTRANSROT
[63]:
COMTRANSROT
# NSCM
1000
END
COVALENTFORM Block¶
This block is needed to define which functional form we will use for bond-stretching (NTBBH), bond-angle bending (NTBAH) and for torsional dihedral (NTBDN). We just use the default options for all functional forms.
[64]:
help(md_imd_file.COVALENTFORM)
Help on COVALENTFORM in module pygromos.files.blocks.imd_blocks object:
class COVALENTFORM(_generic_imd_block)
| COVALENTFORM(NTBBH: int = 0, NTBAH: int = 0, NTBDN: int = 0, content: List[str] = None)
|
| COVALENTFORM Block
|
| The COVALENTFORM Block manages the functional form of the Bonded contributions to the force field.
| It is optional in an imd file to define the block.
|
| Attributes
| ----------
| NTBBH: int
| 0,1 controls bond-stretching potential
| 0: quartic form (default)
| 1: harmonic form
| NTBAH: int
| 0,1 controls bond-angle bending potential
| 0: cosine-harmonic (default)
| 1: harmonic
| NTBDN: int
| 0,1 controls torsional dihedral potential
| 0: arbitrary phase shifts (default)
| 1: phase shifts limited to 0 and 180 degrees.
|
| Method resolution order:
| COVALENTFORM
| _generic_imd_block
| pygromos.files.blocks._general_blocks._generic_gromos_block
| builtins.object
|
| Methods defined here:
|
| __init__(self, NTBBH: int = 0, NTBAH: int = 0, NTBDN: int = 0, content: List[str] = None)
| Initialize self. See help(type(self)) for accurate signature.
|
| ----------------------------------------------------------------------
| Data and other attributes defined here:
|
| __annotations__ = {'NTBAH': <class 'int'>, 'NTBBH': <class 'int'>, 'NT...
|
| name = 'COVALENTFORM'
|
| ----------------------------------------------------------------------
| Methods inherited from _generic_imd_block:
|
| block_to_string(self) -> str
|
| read_content_from_str(self, content: List[str])
|
| ----------------------------------------------------------------------
| Methods inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __copy__(self)
|
| __deepcopy__(self, memo)
|
| __eq__(self, _generic_gromos_block__o: object) -> bool
| Return self==value.
|
| __iter__(self)
|
| __repr__(self)
| Return repr(self).
|
| __str__(self)
| Return str(self).
|
| get_name(self)
|
| ----------------------------------------------------------------------
| Data descriptors inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
|
| ----------------------------------------------------------------------
| Data and other attributes inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __hash__ = None
[65]:
md_imd_file.COVALENTFORM
[65]:
COVALENTFORM
# NTBBH NTBAH NTBDN
0 0 0
END
WRITETRAJ Block¶
Gromos MD simulations produce a massive amount of data and it is impossible to store all the data it produces. The WRITETRAJ block meets this demand: Here you specify how often the coordinate trajectory (NTWX), the velocity trajectory (NTWV), the force trajectory (NTWF), the energy trajectory (NTWE), the free energy trajectory (NTWG) and the block averaged energy trajectory (NTWB) are written out. In the present case, we are only interested in the coordinates (NTWX) and energies (NTWE) and we write them every 100th step. The second switch (NTWSE) defines selection criterion for trajectories: if NTWSE = 0 the normal coordinate trajectory will be written, or if NTWSE \(> 0\) a minimum energy trajectory will be written.
Warning: It makes no sense to write out configurations too often. First, it needs a lot of disk space. Second, the data is highly correlated and so no additional information is gained from it ***
[66]:
help(md_imd_file.WRITETRAJ)
Help on WRITETRAJ in module pygromos.files.blocks.imd_blocks object:
class WRITETRAJ(_generic_imd_block)
| WRITETRAJ(NTWX: int = 0, NTWSE: int = 0, NTWV: int = 0, NTWF: int = 0, NTWE: int = 0, NTWG: int = 0, NTWB: int = 0, content: List[str] = None)
|
| WRITETRAJ
|
| The WRITETRAJ Block manages the output frequency of GROMOS. Here you can determine how often a file should be written.
|
| Attributes
| ----------
| NTWX: int
| controls writing of coordinate trajectory
| 0: no coordinate trajectory is written (default)
| >0: write solute and solvent coordinates every NTWX steps
| <0: write solute coordinates every |NTWX| steps
| NTWSE: int
| >= 0 selection criteria for coordinate trajectory writing
| 0: write normal coordinate trajectory
| >0: write minimum-energy coordinate and energy trajectory (based on the
| energy entry selected by NTWSE and as blocks of length NTWX)
| (see configuration/energy.cc or ene_ana library for indices)
| NTWV: int
| controls writing of velocity trajectory
| 0: no velocity trajectory is written (default)
| >0: write solute and solvent velocities every NTWV steps
| <0: write solute velocities every |NTWV| steps
| NTWF: int
| controls writing of force trajectory
| 0: no force trajectory is written (default)
| >0: write solute and solvent forces every NTWF steps
| <0: write solute forces every |NTWF| steps
| NTWE: int
| >= 0 controls writing of energy trajectory
| 0: no energy trajectory is written (default)
| >0: write energy trajectory every NTWE steps
| NTWG: int
| >= 0 controls writing of free energy trajectory
| 0: no free energy trajectory is written (default)
| >0: write free energy trajectory every NTWG steps
| NTWB: int
| >= 0 controls writing of block-averaged energy trajectory
| 0: no block averaged energy trajectory is written (default)
| >0: write block-averaged energy variables every |NTWB| steps
| (and free energies if NTWG > 0) trajectory
|
| Method resolution order:
| WRITETRAJ
| _generic_imd_block
| pygromos.files.blocks._general_blocks._generic_gromos_block
| builtins.object
|
| Methods defined here:
|
| __init__(self, NTWX: int = 0, NTWSE: int = 0, NTWV: int = 0, NTWF: int = 0, NTWE: int = 0, NTWG: int = 0, NTWB: int = 0, content: List[str] = None)
| Args:
| NTWX:
| NTWSE:
| NTWV:
| NTWF:
| NTWE:
| NTWG:
| NTWB:
|
| ----------------------------------------------------------------------
| Data and other attributes defined here:
|
| __annotations__ = {'NTWB': <class 'int'>, 'NTWE': <class 'int'>, 'NTWF...
|
| __slotnames__ = []
|
| name = 'WRITETRAJ'
|
| ----------------------------------------------------------------------
| Methods inherited from _generic_imd_block:
|
| block_to_string(self) -> str
|
| read_content_from_str(self, content: List[str])
|
| ----------------------------------------------------------------------
| Methods inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __copy__(self)
|
| __deepcopy__(self, memo)
|
| __eq__(self, _generic_gromos_block__o: object) -> bool
| Return self==value.
|
| __iter__(self)
|
| __repr__(self)
| Return repr(self).
|
| __str__(self)
| Return str(self).
|
| get_name(self)
|
| ----------------------------------------------------------------------
| Data descriptors inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
|
| ----------------------------------------------------------------------
| Data and other attributes inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __hash__ = None
[67]:
md_imd_file.WRITETRAJ
[67]:
WRITETRAJ
# NTWX NTWSE NTWV NTWF NTWE NTWG NTWB
10 0 0 0 10 0 0
END
PRINTOUT Block¶
This block is very similar to the WRITETRAJ block but the information about the energies (NTPR) is printed to the output file (.omd). By giving NTPP, dihedral angle transitions are written to the special trajectory (.trs).
[68]:
help(md_imd_file.PRINTOUT)
Help on PRINTOUT in module pygromos.files.blocks.imd_blocks object:
class PRINTOUT(_generic_imd_block)
| PRINTOUT(NTPR: int = 0, NTPP: int = 0, content: List[str] = None)
|
| PRINTOUT block
|
| This Block manages the output frequency into the .omd/std-out file.
|
| Attributes
| ----------
| NTPR: int
| print out energies, etc. every NTPR steps
| NTPP: int
| =1 perform dihedral angle transition monitoring
|
| Method resolution order:
| PRINTOUT
| _generic_imd_block
| pygromos.files.blocks._general_blocks._generic_gromos_block
| builtins.object
|
| Methods defined here:
|
| __init__(self, NTPR: int = 0, NTPP: int = 0, content: List[str] = None)
| Initialize self. See help(type(self)) for accurate signature.
|
| ----------------------------------------------------------------------
| Data and other attributes defined here:
|
| __annotations__ = {'NTPP': <class 'int'>, 'NTPR': <class 'int'>, 'name...
|
| __slotnames__ = []
|
| name = 'PRINTOUT'
|
| ----------------------------------------------------------------------
| Methods inherited from _generic_imd_block:
|
| block_to_string(self) -> str
|
| read_content_from_str(self, content: List[str])
|
| ----------------------------------------------------------------------
| Methods inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __copy__(self)
|
| __deepcopy__(self, memo)
|
| __eq__(self, _generic_gromos_block__o: object) -> bool
| Return self==value.
|
| __iter__(self)
|
| __repr__(self)
| Return repr(self).
|
| __str__(self)
| Return str(self).
|
| get_name(self)
|
| ----------------------------------------------------------------------
| Data descriptors inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
|
| ----------------------------------------------------------------------
| Data and other attributes inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __hash__ = None
[69]:
md_imd_file.PRINTOUT
[69]:
PRINTOUT
# NTPR NTPP
250 0
END
PAIRLIST Block¶
MD++ knows different algorithms for the generation of the pairlist, a list containing the atoms interacting with each other. Here, we use a grid based pairlist generation: the space is discretized into grid cells and only the neighboring cells are searched for interacting partners. The use of this algorithm results in a The pairlist is generated every 5th (NSNB) step. RCUTP and RCUTL are the cutoffs for the pairlist construction for the short-range and the long-range interactions. The pairlist is generated every 5th (NSNB) step. RCUTP and RCUTL are the cutoffs for the pairlist construction for the short-range and the long-range interactions.
[70]:
help(md_imd_file.PAIRLIST)
Help on PAIRLIST in module pygromos.files.blocks.imd_blocks object:
class PAIRLIST(_generic_imd_block)
| PAIRLIST(ALGORITHM: int = 0, NSNB: int = 0, RCUTP: float = 0, RCUTL: float = 0, SIZE: Union[str, float] = 0, TYPE: Union[str, bool] = False, content: List[str] = None)
|
| PAIRLIST Block
|
| This block is controlling the pairlist control.
|
| Attributes
| ----------
| ALGORITHM: int
| standard(0) (gromos96 like pairlist)
| grid(1) (md++ grid pairlist)
| grid_cell(2) (creates a mask)
| NSNB: int
| frequency (number of steps) a pairlist is constructed
| RCUTPL: float
| short-range cut-off in twin-range
| RCUTL: float
| intermediate-range cut-off in twin-range
| SIZE: str, float
| grid cell size (or auto = 0.5 * RCUTP)
| TYPE: str, bool
| chargegoup(0) (chargegroup based cutoff)
| atomic(1) (atom based cutoff)
|
| Method resolution order:
| PAIRLIST
| _generic_imd_block
| pygromos.files.blocks._general_blocks._generic_gromos_block
| builtins.object
|
| Methods defined here:
|
| __init__(self, ALGORITHM: int = 0, NSNB: int = 0, RCUTP: float = 0, RCUTL: float = 0, SIZE: Union[str, float] = 0, TYPE: Union[str, bool] = False, content: List[str] = None)
| Args:
| ALGORITHM:
| NSNB:
| RCUTP:
| RCUTL:
| SIZE:
| TYPE:
|
| ----------------------------------------------------------------------
| Data and other attributes defined here:
|
| __annotations__ = {'ALGORITHM': <class 'int'>, 'NSNB': <class 'int'>, ...
|
| __slotnames__ = []
|
| name = 'PAIRLIST'
|
| ----------------------------------------------------------------------
| Methods inherited from _generic_imd_block:
|
| block_to_string(self) -> str
|
| read_content_from_str(self, content: List[str])
|
| ----------------------------------------------------------------------
| Methods inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __copy__(self)
|
| __deepcopy__(self, memo)
|
| __eq__(self, _generic_gromos_block__o: object) -> bool
| Return self==value.
|
| __iter__(self)
|
| __repr__(self)
| Return repr(self).
|
| __str__(self)
| Return str(self).
|
| get_name(self)
|
| ----------------------------------------------------------------------
| Data descriptors inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
|
| ----------------------------------------------------------------------
| Data and other attributes inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __hash__ = None
[71]:
md_imd_file.PAIRLIST
[71]:
PAIRLIST
# ALGORITHM NSNB RCUTP RCUTL SIZE TYPE
1 5 0.800000 1.400000 0.4 0
END
POSITIONRES Block¶
Finally, we want to restrain the position of our solute. The restraining is achieved by a harmonic special force term with a force constant of CPOR.
[72]:
from pygromos.files.blocks.imd_blocks import POSITIONRES
help(POSITIONRES)
Help on class POSITIONRES in module pygromos.files.blocks.imd_blocks:
class POSITIONRES(_generic_imd_block)
| POSITIONRES(NTPOR: int = 0, NTPORB: int = 0, NTPORS: int = 0, CPOR: int = 0, content: List[str] = None)
|
| POSITIONRES block
|
| This block allows the managment of the Position Restraints during the Simulation.
|
| Attributes
| ----------
| NTPOR:
| 0..3 controls atom positions re(con)straining.
| 0: no position re(con)straints (default)
| 1: restraining with force constant CPOR
| 2: restraining with force constant CPOR weighted by atomic B-factors
| 3: constraining
| NTPORB:
| 0,1 controls reading of reference positions and B-factors
| 0: read reference positions from startup file.
| 1: read reference positions and B-factors from special file
| NTPORS:
| 0,1 controls scaling of reference positions upon pressure scaling
| 0: do not scale reference positions
| 1: scale reference positions
| CPOR:
| >= 0 position restraining force constant
|
| Method resolution order:
| POSITIONRES
| _generic_imd_block
| pygromos.files.blocks._general_blocks._generic_gromos_block
| builtins.object
|
| Methods defined here:
|
| __init__(self, NTPOR: int = 0, NTPORB: int = 0, NTPORS: int = 0, CPOR: int = 0, content: List[str] = None)
| Initialize self. See help(type(self)) for accurate signature.
|
| ----------------------------------------------------------------------
| Data and other attributes defined here:
|
| __annotations__ = {'CPOR': <class 'int'>, 'NTPOR': <class 'int'>, 'NTP...
|
| name = 'POSITIONRES'
|
| ----------------------------------------------------------------------
| Methods inherited from _generic_imd_block:
|
| block_to_string(self) -> str
|
| read_content_from_str(self, content: List[str])
|
| ----------------------------------------------------------------------
| Methods inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __copy__(self)
|
| __deepcopy__(self, memo)
|
| __eq__(self, _generic_gromos_block__o: object) -> bool
| Return self==value.
|
| __iter__(self)
|
| __repr__(self)
| Return repr(self).
|
| __str__(self)
| Return str(self).
|
| get_name(self)
|
| ----------------------------------------------------------------------
| Data descriptors inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
|
| ----------------------------------------------------------------------
| Data and other attributes inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __hash__ = None
[73]:
md_imd_file.add_block(block=POSITIONRES(NTPOR=1, NTPORB=1, CPOR=25000))
md_imd_file.POSITIONRES
[73]:
POSITIONRES
# NTPOR NTPORB NTPORS CPOR
1 1 0 25000
END
Now you should know the main blocks of the Gromos input files.
Perform Thermalisation¶
Next we will run multiple simulations, which will slowly heat up the simulation system. This can easily be done by modifying parameters on the run. First we will setup a gromos system and prepare it. Next we will perform the simulations.
Warning: Depending on your system’s speed this will take up to five hours. As we are using the simulation directory as the working directory there will be an error message after every job which can be ignored. ***
Hint : Have a look at all the output files eq peptide .omd. If anything goes wrong, a message will be printed to the output file.**
[74]:
from pygromos.simulations.modules.preset_simulation_modules import md
## Preparing emin gromos system
in_eq_system = out_eminBox_system.copy()
in_eq_system.name = "eq_thermalisation"
in_eq_system.work_folder = project_dir
### Build position restraints
restrain_res = [k for k in in_eq_system.cnf.residues if(not k in ("CL-", "SOLV"))]
in_eq_system.generate_posres(residues=restrain_res)
### Check simulation params
in_eq_system.imd = md_imd_file
### Set simulation lengths:
in_eq_system.imd.STEP.NSTLIM = 1000 # each temperature will do 1000 steps, this might take some time!
in_eq_system.prepare_for_simulation(not_ligand_residues=["CL-"])
sys_same = in_eq_system.name
in_eq_system
[74]:
GROMOS SYSTEM: eq_thermalisation
################################################################################
WORKDIR: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System
LAST CHECKPOINT: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/emin_box_result.obj
GromosXX_bin: None
GromosPP_bin: None
FILES:
imd: /home/mlehner/PyGromosTools/pygromos/data/simulation_parameters_templates/md_tut.imd
top: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/eq_thermalisation.top
cnf: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/eq_thermalisation.cnf
posres: None
refpos: None
FUTURE PROMISE: False
SYSTEM:
PROTEIN: protein nresidues: 5 natoms: 71
LIGANDS: [] resID: [] natoms: 0
Non-LIGANDS: ['CL-'] nmolecules: 0 natoms: 2
SOLVENT: SOLV nmolecules: 930 natoms: 2790
After preparing the system we can now start simulating.
[75]:
## run Thermalisation.
from pygromos.simulations.modules.preset_simulation_modules import simulation
temperatures = [60, 120, 180, 240, 300] #the temperatures we want to simulate.
print("Heating upt to: ", end="\t")
for runID, temperature in enumerate(temperatures):
print(temperature, end="\t")
#adapt temperature
in_eq_system.imd.MULTIBATH.TEMP0 = [temperature for x in range(in_eq_system.imd.MULTIBATH.NBATHS)]
#turn off the posres for the last run.
if(runID+1 == len(temperatures)): #Last Run
in_eq_system.imd.POSITIONRES.NTPOR = 0
in_eq_system.imd.POSITIONRES.CPOR = 0
out_eq_system = simulation(in_gromos_simulation_system=in_eq_system,
step_name="f_"+sys_same,
simulation_runs=runID+1)
break
else:
out_eq_system = simulation(in_gromos_simulation_system=in_eq_system,
step_name="f_"+sys_same,
simulation_runs=runID+1,
analysis_script=None,
verbose=False)
in_eq_system = out_eq_system
print("done")
Heating upt to: 60 120 180 240 300 ################################################################################
f_eq_thermalisation
################################################################################
Script: /home/mlehner/PyGromosTools/pygromos/simulations/hpc_queuing/job_scheduling/schedulers/simulation_scheduler.py
################################################################################
Simulation Setup:
################################################################################
steps_per_run: 1000
equis: 0
simulation runs: 5
################################################################################
submit final analysis part
/home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/f_eq_thermalisation/ana_out.log
/home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/f_eq_thermalisation/job_analysis.py
ANA jobID: 0
done
[76]:
#This took some time, so lets store it! So we can use it later again.
out_eq_therm_system_path = out_eq_system.save(out_eq_system.work_folder+"/eq_therm_result2.obj")
Analysis of Thermalisation:¶
Next we want to check the results of our Thermalisation. We will have a look at the coordinate trajectory and the temperatures of the system.
Check the coordinate traj¶
First we will check the trajectory of the termalisation. Again we first need to shift the coordinates, so we can se the fully connected peptide.
[77]:
coordinate_traj = out_eq_system.trc
coordinate_traj.image_molecules()
view = coordinate_traj.view
view
Check the temperature¶
After this, we would like to see the temperature development. First lets collect all temperature calculations from the simulation:
[78]:
energy_traj = out_eq_system.tre
temperatures= energy_traj.get_temperature()
temperatures
[78]:
| bath1 | bath2 | |
|---|---|---|
| time | ||
| 0.00 | 339.434523 | 378.651407 |
| 0.02 | 140.118831 | 389.835477 |
| 0.04 | 160.978927 | 383.542770 |
| 0.06 | 202.329896 | 379.484185 |
| 0.08 | 151.866737 | 349.959059 |
| ... | ... | ... |
| 9.90 | 335.579171 | 309.355612 |
| 9.92 | 275.800192 | 310.621616 |
| 9.94 | 265.092556 | 307.698791 |
| 9.96 | 300.889369 | 300.488861 |
| 9.98 | 318.342788 | 305.876061 |
500 rows × 2 columns
Next we want to visualize the data to get an impresstionof what happened. Can you see how the Temperature was constantly raised per simulation and started to equilibrate forming plateus?
[79]:
from matplotlib import pyplot as plt
plt.plot(temperatures.bath1, label="TBath1")
plt.plot(temperatures.bath2, label="TBath2")
plt.legend()
plt.ylabel("$T~[K]$")
plt.xlabel("$t~[ps]$")
[79]:
Text(0.5, 0, '$t~[ps]$')
Molecular dynamics sampling simulation.¶
The equilibration period already produced a short simulation at constant temperature and volume. At this point we want to elongate the simulation to a nanosecond under constant temperature and pressure.
Simulation Paramters¶
First, we don’t use position restraining anymore and so, the POSITIONRES block in the next step. Next we want to simulate under constant pressure rather than constant volume (NVT-> NPT). For this purpose we have to add an additional block:
[80]:
from pygromos.data.simulation_parameters_templates import template_md_tut as template_md
from pygromos.files.simulation_parameters.imd import Imd
imd_file = Imd(template_md)
help(imd_file.PRESSURESCALE)
Help on PRESSURESCALE in module pygromos.files.blocks.imd_blocks object:
class PRESSURESCALE(_generic_imd_block)
| PRESSURESCALE(COUPLE: int = 0, SCALE: int = 0, COMP: float = 0, TAUP: float = 0, VIRIAL: int = 0, SEMIANISOTROPIC: List[int] = [], PRES0: List[List[float]] = [[0, 0, 0], [0, 0, 0], [0, 0, 0]], content: List[str] = None)
|
| PRESSURESCALE Block
| This block controls the barostat of the simulation
|
| Attributes
| -----------
| COUPLE: int
| off(0), calc(1), scale(2)
| SCALE: int
| off(0), iso(1), aniso(2), full(3), semianiso(4)
| COMP: float
| compessibility
| TAUP: float
| coupling strength
| VIRIAL: int
| none(0), atomic(1), group(2)
| SEMIANISOTROPIC: List[int]
| (semianisotropic couplings: X, Y, Z)
| e.g. 1 1 2: x and y jointly coupled and z separately coupled
| e.g. 0 0 1: constant area (xy-plane) and z coupled to a bath
| PRES0: List[List[float]]
| reference pressure
|
| Method resolution order:
| PRESSURESCALE
| _generic_imd_block
| pygromos.files.blocks._general_blocks._generic_gromos_block
| builtins.object
|
| Methods defined here:
|
| __init__(self, COUPLE: int = 0, SCALE: int = 0, COMP: float = 0, TAUP: float = 0, VIRIAL: int = 0, SEMIANISOTROPIC: List[int] = [], PRES0: List[List[float]] = [[0, 0, 0], [0, 0, 0], [0, 0, 0]], content: List[str] = None)
| Initialize self. See help(type(self)) for accurate signature.
|
| read_content_from_str(self, content: List[str])
|
| ----------------------------------------------------------------------
| Data and other attributes defined here:
|
| __annotations__ = {'COMP': <class 'float'>, 'COUPLE': <class 'int'>, '...
|
| __slotnames__ = []
|
| name = 'PRESSURESCALE'
|
| ----------------------------------------------------------------------
| Methods inherited from _generic_imd_block:
|
| block_to_string(self) -> str
|
| ----------------------------------------------------------------------
| Methods inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __copy__(self)
|
| __deepcopy__(self, memo)
|
| __eq__(self, _generic_gromos_block__o: object) -> bool
| Return self==value.
|
| __iter__(self)
|
| __repr__(self)
| Return repr(self).
|
| __str__(self)
| Return str(self).
|
| get_name(self)
|
| ----------------------------------------------------------------------
| Data descriptors inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
|
| ----------------------------------------------------------------------
| Data and other attributes inherited from pygromos.files.blocks._general_blocks._generic_gromos_block:
|
| __hash__ = None
In the PRESSURESCALE block we tell Gromos to calculate and scale the pressure by setting COUPLE to \(2\). As the box should be isotropically scaled we set SCALE equal to \(1\). The weak-coupling method (Sec. 2-12.2.2) uses two additional parameters: COMP is the isothermal compressibility and TAUP is the coupling time. We are calculating the molecular virial (VIRIAL is equal to \(2\)), so intramolecular forces don’t contribute to the pressure. The next line is only used for semi-anisotropic pressure coupling and can be ignored in our case. Finally, we have to specify the reference pressure in a tensor form
[81]:
imd_file.PRESSURESCALE
[81]:
PRESSURESCALE
# COUPLE SCALE COMP TAUP VIRIAL
2 1 0.000458 0.500000 2
# SEMIANISOTROPIC COUPLINGS(X, Y, Z)
1 1 1
# PRES0(1...3,1...3)
0.06102 0.0 0.0
0.0 0.06102 0.0
0.0 0.0 0.06102
END
Perform MD - Production¶
In the other blocks only minor things have changed: the temperature was set to 300K and the trajectories are written out less often (every 250th step only). Next we are going to set up the Gromos System. We will remove the position restraints and the old trajectories.
[82]:
from pygromos.simulations.modules.preset_simulation_modules import md
in_md_system = out_eq_system.copy()
in_md_system.work_folder = project_dir
in_md_system.name = "md"
### Check simulation params
in_md_system.imd = imd_file
## imd parameters
in_md_system.imd.STEP.NSTLIM = 1000
in_md_system.imd.WRITETRAJ.NTWX = 10
in_md_system.imd.WRITETRAJ.NTWE = 10
in_md_system.imd.INITIALISE.NTIVEL = 0
in_md_system.prepare_for_simulation()
in_md_system
[82]:
GROMOS SYSTEM: md
################################################################################
WORKDIR: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System
LAST CHECKPOINT: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/eq_therm_result2.obj
GromosXX_bin: None
GromosPP_bin: None
FILES:
imd: /home/mlehner/PyGromosTools/pygromos/data/simulation_parameters_templates/md_tut.imd
top: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/f_eq_thermalisation.top
cnf: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/f_eq_thermalisation.cnf
posres: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/f_eq_thermalisation.pos
refpos: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/f_eq_thermalisation.rpf
FUTURE PROMISE: False
SYSTEM:
PROTEIN: protein nresidues: 5 natoms: 71
LIGANDS: ['CL6', 'CL7'] resID: [6, 7] natoms: 2
Non-LIGANDS: [] nmolecules: 0 natoms: 0
SOLVENT: SOLV nmolecules: 930 natoms: 2790
[83]:
out_md_system = md(in_gromos_system=in_md_system,
step_name="g_"+in_md_system.name,
simulation_runs=3)
out_md_system
################################################################################
g_md
################################################################################
Script: /home/mlehner/PyGromosTools/pygromos/simulations/hpc_queuing/job_scheduling/schedulers/simulation_scheduler.py
################################################################################
Simulation Setup:
################################################################################
steps_per_run: 1000
equis: 0
simulation runs: 3
################################################################################
submit final analysis part
/home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/g_md/ana_out.log
/home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/g_md/job_analysis.py
ANA jobID: 0
[83]:
GROMOS SYSTEM: g_md
################################################################################
WORKDIR: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System
LAST CHECKPOINT: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/eq_therm_result2.obj
GromosXX_bin: None
GromosPP_bin: None
FILES:
imd: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/g_md/input/g_md.imd
top: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/g_md/input/g_md.top
cnf: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/g_md/analysis/data/g_md.cnf
posres: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/g_md/input/g_md.pos
refpos: /home/mlehner/PyGromosTools/docs/sphinx_project/Tutorials/example_files/Tutorial_System/g_md/input/g_md.rpf
FUTURE PROMISE: True
SYSTEM:
PROTEIN: protein nresidues: 5 natoms: 71
LIGANDS: ['CL6', 'CL7'] resID: [6, 7] natoms: 2
Non-LIGANDS: [] nmolecules: 0 natoms: 0
SOLVENT: SOLV nmolecules: 930 natoms: 2790
[84]:
out_md_system_path = out_md_system.save(out_md_system.work_folder+"/md_result.obj")
After all the jobs are finished, you should start to analyse the trajectories.
Analysis¶
First Look Analysis¶
Let’s first have a look on the final coordinates.
[85]:
out_md_system.cnf.shift_periodic_boundary()
out_md_system.cnf.recenter_pbc()
out_md_system.cnf.recreate_view()
out_md_system.cnf.view
[86]:
coordinate_traj = out_md_system.trc
coordinate_traj.image_molecules()
coordinate_traj.recreate_view()
view = coordinate_traj.view
view
/home/mlehner/anaconda3/envs/pygro2/lib/python3.9/site-packages/mdtraj/core/trajectory.py:438: UserWarning: top= kwargs ignored since this file parser does not support it
warnings.warn('top= kwargs ignored since this file parser does not support it')
Energy Analysis¶
The program calculates the average of the specified properties as well as the root-mean-square deviations (rmsd) and a statistical error estimate (error est.). The error estimate is calculated from block averages of growing sizes extrapolating to infinite block size1
Warning: Sometimes the error estimates are NaN (not a number), which is due to the fact that we do not have enough values to calculate a meaningful error estimate. ***
[87]:
energy_traj = out_md_system.tre
total_energies = energy_traj.get_totals()
total_energies
[87]:
| totene | totkin | totpot | totcov | totbond | totangle | totimproper | totdihedral | totcrossdihedral | totnonbonded | ... | eds_vmix | eds_vr | eds_emax | eds_emin | eds_globmin | eds_globminfluc | entropy | totqm | totbsleus | totrdc | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| time | |||||||||||||||||||||
| 0.00 | -47298.79052 | 1618.643530 | -48917.43405 | 252.617847 | 0.0 | 120.249867 | 38.645933 | 93.722048 | 0.0 | -49170.05190 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 0.02 | -46269.03164 | 2464.331707 | -48733.36335 | 221.036528 | 0.0 | 89.627692 | 45.038378 | 86.370457 | 0.0 | -48954.39987 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 0.04 | -45353.28866 | 3010.497757 | -48363.78641 | 279.233662 | 0.0 | 123.663886 | 51.890773 | 103.679003 | 0.0 | -48643.02008 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 0.06 | -44504.63121 | 3341.937138 | -47846.56835 | 292.535309 | 0.0 | 139.711508 | 45.975944 | 106.847857 | 0.0 | -48139.10366 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 0.08 | -43753.16520 | 3684.261609 | -47437.42681 | 288.216128 | 0.0 | 129.742936 | 56.731466 | 101.741727 | 0.0 | -47725.64294 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 5.90 | -33587.58374 | 7335.329137 | -40922.91288 | 229.219891 | 0.0 | 115.319382 | 49.971611 | 63.928898 | 0.0 | -41152.13277 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 5.92 | -33587.32408 | 7184.037953 | -40771.36203 | 224.153707 | 0.0 | 90.114185 | 48.815956 | 85.223565 | 0.0 | -40995.51574 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 5.94 | -33542.19033 | 7240.118961 | -40782.30929 | 209.515148 | 0.0 | 100.347480 | 44.342135 | 64.825533 | 0.0 | -40991.82444 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 5.96 | -33532.49241 | 7225.276174 | -40757.76858 | 210.539388 | 0.0 | 103.851765 | 39.627214 | 67.060410 | 0.0 | -40968.30797 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 5.98 | -33536.24937 | 7231.265778 | -40767.51515 | 236.970723 | 0.0 | 98.997937 | 58.265716 | 79.707070 | 0.0 | -41004.48587 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
300 rows × 43 columns
[88]:
#Next we plot some data of the energies to se their development.
time_axis = energy_traj.time
total_energies.totpot.plot(legend=True)
total_energies.totkin.plot(legend=True)
total_energies.totene.plot(xlabel="t [ps]", ylabel="V [kJ/mol]", title="Simulation Data", legend=True)
[88]:
<AxesSubplot:title={'center':'Simulation Data'}, xlabel='t [ps]', ylabel='V [kJ/mol]'>
In the Following we want to see the different contributions of the molecules with their VdW and Culomb interaxtions. we add up the van der Waals and Coulomb energies of the peptide-peptide interactions.
Now we want to look at two variables: * peptide_water_nonbonded * peptide_Cl_nonbonded
The information about the nonbondeds can be retrieved from the ForceGroup Nonbonded contributions. If you check the used imd file, you reckognize three Force groups in the FORCEGROUP block. These are limited by the last atom of the group and you can check the group atoms in the topology or coordinate file. They should correspond here to: 1. Peptide 2. 2 Cl- ions 3. Water molecules
The interactions of these forcegroups are stored in the following pattern, which can be adapted to any force group number (PseudoCode: for i in range(1+nFroceGroups) for j in range(i, nFroceGroups)):
ForceGroup - 1. ForceGroup: intra force group nonbondeds (peptide - peptide)
ForceGroup - 2. ForceGroup: inter force group nonbondeds between force group 1 and 2 (peptide - Cl-)
ForceGroup - 3. ForceGroup: inter force group nonbondeds between force group 1 and 3 (peptide - Water)
ForceGroup - 2. ForceGroup: intra force group nonbondeds (Cl- - Cl-)
ForceGroup - 3. ForceGroup: inter force group nonbondeds between force group 2 and 3 (Cl- - Water)
ForceGroup - 3. ForceGroup: intra force group nonbondeds (Water - Water)
[89]:
#First lets get the Force Group Energy contributions:
forceGroupNonbondedContributions = energy_traj.get_nonbondedContributions()
#Here we give each Force Group contribution category nice names:
peptide_peptide_nonbonded = forceGroupNonbondedContributions[1][1]
peptide_Cl_nonbonded = forceGroupNonbondedContributions[1][2]
peptide_water_nonbonded = forceGroupNonbondedContributions[1][3]
Cl_Cl_nonbonded = forceGroupNonbondedContributions[2][2]
Cl_water_nonbonded = forceGroupNonbondedContributions[2][3]
water_water_nonbonded = forceGroupNonbondedContributions[3][3]
[90]:
#Here we look at the different Nonbonded contributions of each
peptide_peptide_nonbonded.plot(title="Peptide_intra_Nonbondeds", ylabel="V [kJ/mol]", xlabel="writeOuts", subplots=True)
Cl_Cl_nonbonded.plot(title="CL_intra_Nonbondeds", ylabel="V [kJ/mol]", xlabel="writeOuts", subplots=True)
water_water_nonbonded.plot(title="Water_intra_Nonbondeds", ylabel="V [kJ/mol]", xlabel="writeOuts", subplots=True)
[90]:
array([<AxesSubplot:xlabel='writeOuts', ylabel='V [kJ/mol]'>,
<AxesSubplot:xlabel='writeOuts', ylabel='V [kJ/mol]'>,
<AxesSubplot:xlabel='writeOuts', ylabel='V [kJ/mol]'>,
<AxesSubplot:xlabel='writeOuts', ylabel='V [kJ/mol]'>],
dtype=object)
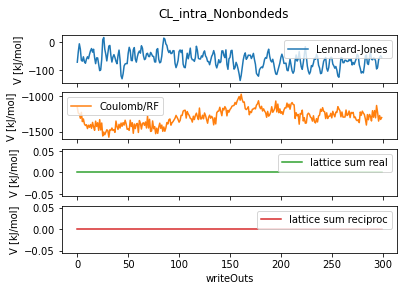
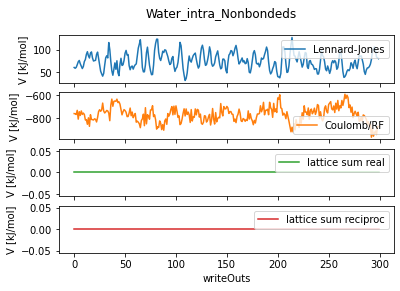
[91]:
#Next we calculate the sum of all contributions
#The sum over the axis 1 sums up all contribution types for a timestep t.
peptide_peptide_nonbonded.sum(axis=1).plot(label="peptide - peptide", legend=True)
peptide_Cl_nonbonded.sum(axis=1).plot(label="peptide - Cl-", legend=True)
peptide_water_nonbonded.sum(axis=1).plot(label="peptide - Water", ylabel="V [kJ/mol]", xlabel="writeOuts", title="Peptide Nonbondeds", legend=True)
[91]:
<AxesSubplot:title={'center':'Peptide Nonbondeds'}, xlabel='writeOuts', ylabel='V [kJ/mol]'>
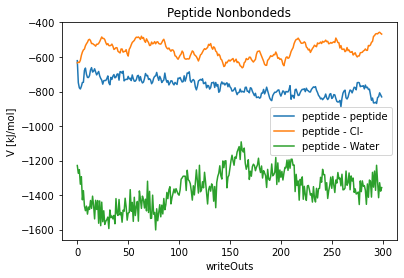
[92]:
# Here we look at the total nonbonded Contributions of the peptide
peptide_nonbonded = peptide_peptide_nonbonded+peptide_Cl_nonbonded+peptide_water_nonbonded
#The sum over the axis 1 sums up all contribution types for a timestep t.
peptide_nonbonded.sum(axis=1).plot( label="peptide", ylabel="V [kJ/mol]", xlabel="steps", title="Total Nonbonded Contributions of peptide", legend=True)
[92]:
<AxesSubplot:title={'center':'Total Nonbonded Contributions of peptide'}, xlabel='steps', ylabel='V [kJ/mol]'>
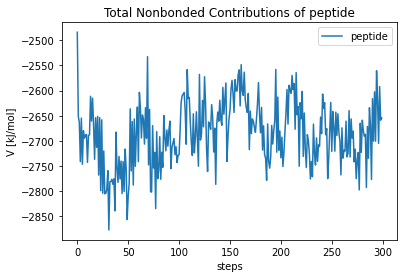
Coordinate Analysis¶
In the next step, we want to have a look at the coordinates of our simulation. first we will dcalculate the rmsd, second we will analyse the distance of the CL- to the positive charged atoms in the peptide.
Warning: Under Development ;) ***
[93]:
final_coordinate = out_md_system.cnf
coordinate_traj = out_md_system.trc
[94]:
coordinate_traj.rmsd(1).sum(axis=1).plot(ylabel="RMSD", xlabel="t [ps]")
[94]:
<AxesSubplot:xlabel='t [ps]', ylabel='RMSD'>
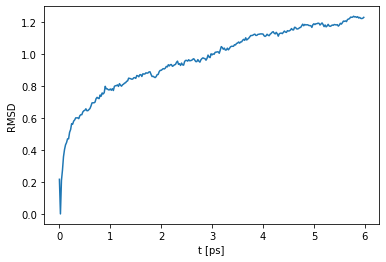
Distance CL- to the peptide¶
[95]:
print("CL- atoms:", [(ap.atomType, ap.atomID) for ap in final_coordinate.POSITION if(ap.resName == "CL-")])
print("ARG atoms:", [(ap.atomType, ap.atomID) for ap in final_coordinate.POSITION if(ap.resName == "ARG")])
print("LYSH atoms:", [(ap.atomType, ap.atomID) for ap in final_coordinate.POSITION if(ap.resName == "LYSH")])
print("NTERM atoms:", [(ap.atomType, ap.atomID) for ap in final_coordinate.POSITION if(ap.resName == "VAL")])
CL- atoms: [('CL', 72), ('CL', 73)]
ARG atoms: [('N', 29), ('H', 30), ('CA', 31), ('CB', 32), ('CG', 33), ('CD', 34), ('NE', 35), ('HE', 36), ('CZ', 37), ('NH1', 38), ('HH11', 39), ('HH12', 40), ('NH2', 41), ('HH21', 42), ('HH22', 43), ('C', 44), ('O', 45)]
LYSH atoms: [('N', 46), ('H', 47), ('CA', 48), ('CB', 49), ('CG', 50), ('CD', 51), ('CE', 52), ('NZ', 53), ('HZ1', 54), ('HZ2', 55), ('HZ3', 56), ('C', 57), ('O', 58)]
NTERM atoms: [('H1', 1), ('H2', 2), ('N', 3), ('H3', 4), ('CA', 5), ('CB', 6), ('CG1', 7), ('CG2', 8), ('C', 9), ('O', 10)]
Calculate the distances now to the different CL- ions.
[96]:
coordinate_traj.distances(atom_pairs=[[72, 37], [73, 37]]).plot(label="CL-1/2 - ARG-CZ", legend=True, xlabel="t [ps]", ylabel="r [nm]")
[96]:
<AxesSubplot:xlabel='t [ps]', ylabel='r [nm]'>
[97]:
coordinate_traj.distances(atom_pairs=[[72, 53], [73, 53]]).plot(label="CL-1/2 - LYSH-NZ", legend=True, xlabel="t [ps]", ylabel="r [nm]")
[97]:
<AxesSubplot:xlabel='t [ps]', ylabel='r [nm]'>
[98]:
coordinate_traj.distances(atom_pairs=[[72, 3], [73, 3]]).plot(label="CL-1/2 - N-TERM", legend=True, xlabel="t [ps]", ylabel="r [nm]")
[98]:
<AxesSubplot:xlabel='t [ps]', ylabel='r [nm]'>
[ ]: